








生物技术通报 ›› 2025, Vol. 41 ›› Issue (2): 270-283.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0537
• 研究报告 • 上一篇
收稿日期:2024-06-04
出版日期:2025-02-26
发布日期:2025-02-28
通讯作者:
张海娥,女,博士,研究员,研究方向 :板栗育种;E-mail: zhang33haie4@163.com作者简介:杨涌,男,硕士研究生,研究方向 :板栗育种;E-mail: 1725722566@qq.com
基金资助:
YANG Yong( ), CAO Rui, KANG Xiao-xiao, LIU Jing, WANG Xuan, ZHANG Hai-e(
), CAO Rui, KANG Xiao-xiao, LIU Jing, WANG Xuan, ZHANG Hai-e( )
)
Received:2024-06-04
Published:2025-02-26
Online:2025-02-28
摘要:
目的 从板栗基因组中鉴定类黄酮合成通路13个基因家族,分析其在果实成熟过程和果实顶部芽组织PCD过程中的表达模式,为后续研究板栗类黄酮相关基因功能奠定基础。 方法 利用生物信息学技术鉴定板栗类黄酮合成通路13个基因家族,对其系统进化关系、蛋白理化性质、蛋白结构、基因结构、顺式作用元件、共线性关系、密码子偏好性等内容进行分析,同时,利用转录组和RT-qPCR分析不同发育时期板栗果实成熟过程和果实顶部芽组织PCD过程表达模式。 结果 板栗类黄酮合成通路13个基因家族57个成员分布在12个染色体及4个contig上。各基因家族包含1-2个关键结构域,各基因家族成员内含子数量具有一定的相似性。顺式作用元件分析结果显示,大量的生长发育相关元件、激素响应元件以及胁迫响应元件存在板栗中类黄酮合成通路基因家族成员启动子上。在板栗基因组内共发现7对共线性基因对,同时,发现4个基因与8个单双子叶植物均有共线性基因。密码子偏好性发现RSCU值大于1的偏好A/U结尾,且ENC值均超过35。转录组与RT-qPCR结果都显示,多个黄酮合成通路基因在板栗果实成熟过程和果实顶部芽组织PCD过程表达量上调。 结论 共鉴定57个板栗类黄酮合成通路基因,CmLAR2、CmCHI1、Cm4CL1、CmC4H3、CmPAL1、CmPAL2、Cm4CL6和CmANR3在板栗果实成熟过程和果实顶部芽组织PCD过程均表达量上调,可能参与板栗果实成熟过程和板栗芽组织PCD过程。
杨涌, 曹蕊, 康肖肖, 刘静, 王旋, 张海娥. 板栗类黄酮合成通路13个基因家族的鉴定及表达分析[J]. 生物技术通报, 2025, 41(2): 270-283.
YANG Yong, CAO Rui, KANG Xiao-xiao, LIU Jing, WANG Xuan, ZHANG Hai-e. Identification and Expression Analysis of 13 Gene Families in the Chestnut Flavonoid Synthesis Pathway[J]. Biotechnology Bulletin, 2025, 41(2): 270-283.
| 基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| CmLAR2 | GTGGAACCAGCCCTGACC | GGTAGGGCCAAGCAGCAA |
| CmCHI1 | AGCGTCTCTTCCGGGAGT | CGCCGAGGAACAGTGTGT |
| Cm4CL1 | TGACTCAGGCCGACGAGA | CGACTTGTTGTGCCACGC |
| CmC4H3 | AGGGTGTGGAGTTCGGGT | ATGCTCGCCGTAAACCGT |
| CmPAL1 | AGGCCCTCCAATGCAACC | CGGCAGTGACCTGAGCAA |
| CmPAL2 | CGACTCGTGCCGCTATGT | CGGAGTGGCAAGCATGGA |
| Cm4CL6 | ATCCAGAGGCCACGGCTA | CTCGGCAGGGGCTACTTG |
| CmANR3 | CACCGTTCGGTCTGACCC | GGGTGCATTGAAACTCTCTGG |
| CmActin | ATTCACGAGACCACCTACA | TGCCACAACCTTAATCTTCAT |
表1 引物序列
Table 1 Primer sequences
| 基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| CmLAR2 | GTGGAACCAGCCCTGACC | GGTAGGGCCAAGCAGCAA |
| CmCHI1 | AGCGTCTCTTCCGGGAGT | CGCCGAGGAACAGTGTGT |
| Cm4CL1 | TGACTCAGGCCGACGAGA | CGACTTGTTGTGCCACGC |
| CmC4H3 | AGGGTGTGGAGTTCGGGT | ATGCTCGCCGTAAACCGT |
| CmPAL1 | AGGCCCTCCAATGCAACC | CGGCAGTGACCTGAGCAA |
| CmPAL2 | CGACTCGTGCCGCTATGT | CGGAGTGGCAAGCATGGA |
| Cm4CL6 | ATCCAGAGGCCACGGCTA | CTCGGCAGGGGCTACTTG |
| CmANR3 | CACCGTTCGGTCTGACCC | GGGTGCATTGAAACTCTCTGG |
| CmActin | ATTCACGAGACCACCTACA | TGCCACAACCTTAATCTTCAT |
| 基因家族 Gene family | 以马可夫模型编号 Pfam ID | 结构域名称 Domain name |
|---|---|---|
| PAL | PF00221 | 芳香族氨基酸裂解酶 Aromatic amino acid lyase |
| 4CL | PF13193, PF00501 | AMP结合酶C末端结构域,AMP结合酶结构域 AMP-binding enzyme C-terminal domain, AMP-binding enzyme domain |
| CHS | PF02797, PF00195 | 查尔酮和二苯乙烯合酶,N/C末端结构域 Chalcone and stilbene synthases, N/C-terminal domain |
| CHI | PF02431 | 查尔酮黄烷酮异构酶 Chalcone-flavanone isomerase |
| LAR | PF05368 | NmrA-like结构域 NmrA-like domain |
| DFR | PF01370 | NAD依赖性差向异构酶结构域 NAD dependent epimerase domain |
| ANR | ||
| F3H | PF03171, PF14226 | 2OG-FeII_Oxy结构域,DIOX_N结构域 2OG-FeII_Oxy domain, DIOX_N domain |
| FLS | ||
| ANS | ||
| C4H | PF00067 | 细胞色素P450 Cytochrome P450 |
| F3′5′H | ||
| FNSII |
表2 13个基因家族成员蛋白结构分析
Table 2 Analysis of the protein structures of the 13 gene family members
| 基因家族 Gene family | 以马可夫模型编号 Pfam ID | 结构域名称 Domain name |
|---|---|---|
| PAL | PF00221 | 芳香族氨基酸裂解酶 Aromatic amino acid lyase |
| 4CL | PF13193, PF00501 | AMP结合酶C末端结构域,AMP结合酶结构域 AMP-binding enzyme C-terminal domain, AMP-binding enzyme domain |
| CHS | PF02797, PF00195 | 查尔酮和二苯乙烯合酶,N/C末端结构域 Chalcone and stilbene synthases, N/C-terminal domain |
| CHI | PF02431 | 查尔酮黄烷酮异构酶 Chalcone-flavanone isomerase |
| LAR | PF05368 | NmrA-like结构域 NmrA-like domain |
| DFR | PF01370 | NAD依赖性差向异构酶结构域 NAD dependent epimerase domain |
| ANR | ||
| F3H | PF03171, PF14226 | 2OG-FeII_Oxy结构域,DIOX_N结构域 2OG-FeII_Oxy domain, DIOX_N domain |
| FLS | ||
| ANS | ||
| C4H | PF00067 | 细胞色素P450 Cytochrome P450 |
| F3′5′H | ||
| FNSII |
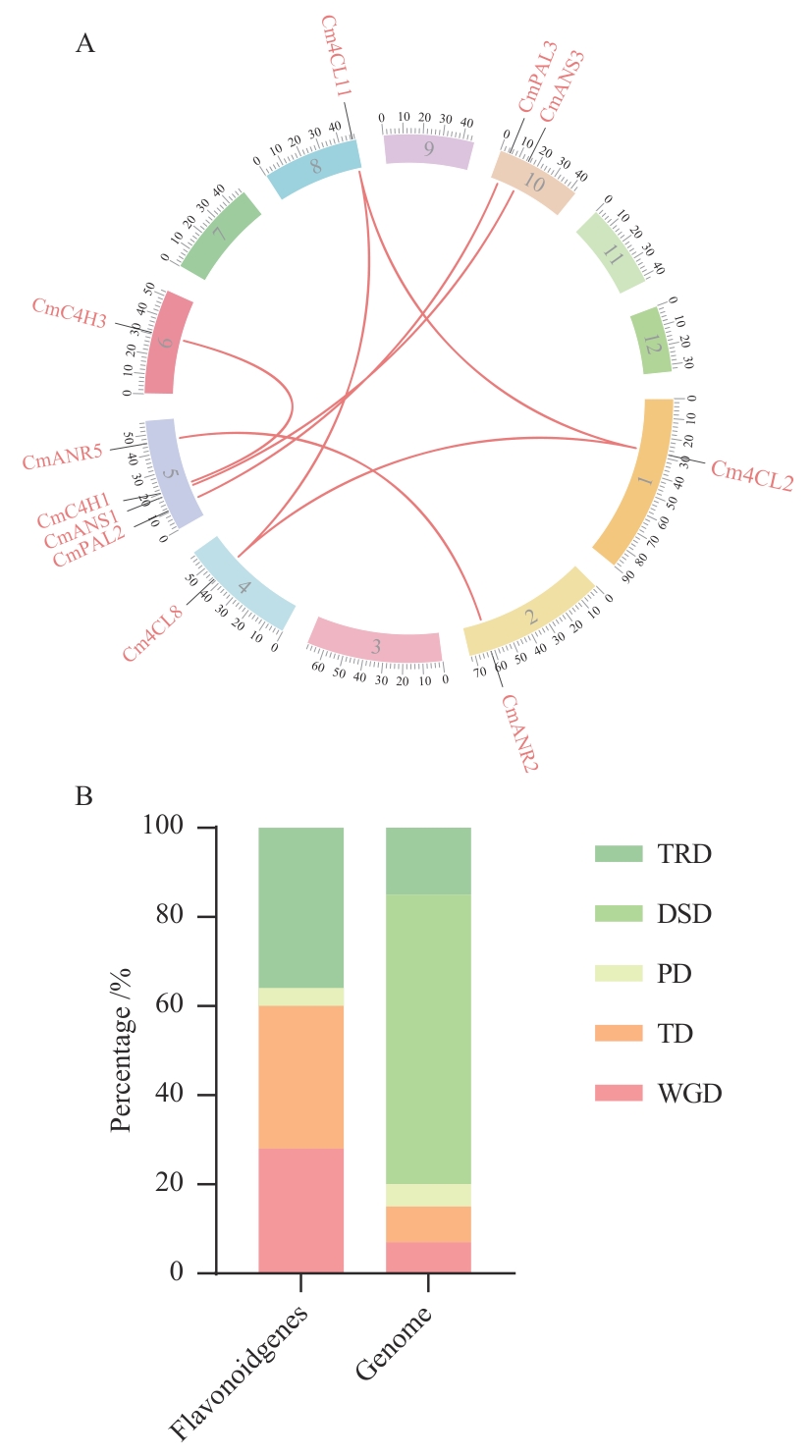
图5 13个基因家族基因组内共线性(A)和基因对复制类型(B)TRD、DSD、PD、TD及WGD代表转座子复制、分散复制、近端复制、串联复制及全基因组复制。下同
Fig. 5 Collinearity within 13 gene families (A) and gene pairs replication types (B)TRD, DSD, PD, TD and WGD indicate transposon duplication, dispersed duplication, proximal duplication, tandem duplication and whole genome duplication. The same below
| 复制类型 Duplication types | 基因对 Gene pairs | 非同义替换率 Ka | 同义替换率 Ks | 非同义替换率/同义替换率 Ka/Ks | |
|---|---|---|---|---|---|
| WGD | CmPAL3 | CmPAL2 | 0.123 822 763 | 1.465 415 048 | 0.084 496 719 |
| WGD | Cm4CL2 | Cm4CL11 | 0.383 354 993 | 1.384 646 007 | 0.276 861 372 |
| WGD | CmANR5 | CmANR2 | 0.549 975 334 | 2.288 010 781 | 0.240 372 702 |
| WGD | CmC4H1 | CmC4H3 | 0.085 827 583 | 1.945 979 279 | 0.044 105 086 |
| WGD | CmANS3 | CmANS1 | 0.219 052 838 | 1.816 044 933 | 0.120 620 825 |
| WGD | Cm4CL8 | Cm4CL2 | 0.366 520 820 | 1.302 665 198 | 0.281 362 257 |
| WGD | Cm4CL8 | Cm4CL11 | 0.372 236 766 | 1.132 735 123 | 0.328 617 661 |
| PD | Cm4CL14 | Cm4CL15 | 0.045 708 825 | 0.165 863 036 | 0.275 581 749 |
| TD | Cm4CL3 | Cm4CL4 | 0.143 933 551 | 0.443 981 508 | 0.324 188 167 |
| TD | CmPAL5 | CmPAL6 | 0.054 029 598 | 0.093 294 174 | 0.579 131 530 |
| TD | CmFNSII2 | CmFNSII3 | 0.139 352 161 | 0.427 416 010 | 0.326 034 023 |
| TD | CmDFR1 | CmDFR2 | 0.189 572 148 | 0.623 062 431 | 0.304 258 672 |
| TD | CmANR3 | CmANR4 | 0.137 094 524 | 0.393 608 899 | 0.348 301 384 |
| TD | Cm4CL4 | Cm4CL5 | 0.133 265 871 | 0.425 828 776 | 0.312 956 470 |
| TD | Cm4CL9 | Cm4CL10 | 0.103 441 680 | 0.273 134 833 | 0.378 720 204 |
| TD | CmPAL4 | CmPAL5 | 0.052 152 683 | 0.128 965 485 | 0.404 392 564 |
| TRD | CmCHS1 | CmCHS2 | 0.644 246 280 | NaN (high sequence divergence) | |
| TRD | CmCHS3 | CmCHS4 | 0.587 247 232 | 2.429 515 767 | 0.241 713 694 |
| TRD | CmLAR2 | CmLAR1 | 0.263 851 247 | 1.291 720 422 | 0.204 263 432 |
| TRD | CmF3'5'H1 | CmF3'5'H2 | 0.090 142 821 | 0.211 897 324 | 0.425 408 022 |
| TRD | CmFLS2 | CmFLS3 | 0.190 856 593 | 1.395 097 547 | 0.136 805 196 |
| TRD | CmANS2 | CmANS1 | 0.636 070 470 | 3.657 827 926 | 0.173 892 945 |
| TRD | CmFLS1 | CmFLS3 | 0.467 506 223 | 3.335 289 988 | 0.140 169 588 |
| TRD | CmANR7 | CmANR6 | 0.015 019 275 | 0.051 683 849 | 0.290 599 013 |
| TRD | Cm4CL6 | Cm4CL1 | 0.148 018 360 | 1.985 685 394 | 0.074 542 705 |
表3 基因对Ka/Ks值的计算
Table 3 Calculations of the gene-pairs Ka/Ks values
| 复制类型 Duplication types | 基因对 Gene pairs | 非同义替换率 Ka | 同义替换率 Ks | 非同义替换率/同义替换率 Ka/Ks | |
|---|---|---|---|---|---|
| WGD | CmPAL3 | CmPAL2 | 0.123 822 763 | 1.465 415 048 | 0.084 496 719 |
| WGD | Cm4CL2 | Cm4CL11 | 0.383 354 993 | 1.384 646 007 | 0.276 861 372 |
| WGD | CmANR5 | CmANR2 | 0.549 975 334 | 2.288 010 781 | 0.240 372 702 |
| WGD | CmC4H1 | CmC4H3 | 0.085 827 583 | 1.945 979 279 | 0.044 105 086 |
| WGD | CmANS3 | CmANS1 | 0.219 052 838 | 1.816 044 933 | 0.120 620 825 |
| WGD | Cm4CL8 | Cm4CL2 | 0.366 520 820 | 1.302 665 198 | 0.281 362 257 |
| WGD | Cm4CL8 | Cm4CL11 | 0.372 236 766 | 1.132 735 123 | 0.328 617 661 |
| PD | Cm4CL14 | Cm4CL15 | 0.045 708 825 | 0.165 863 036 | 0.275 581 749 |
| TD | Cm4CL3 | Cm4CL4 | 0.143 933 551 | 0.443 981 508 | 0.324 188 167 |
| TD | CmPAL5 | CmPAL6 | 0.054 029 598 | 0.093 294 174 | 0.579 131 530 |
| TD | CmFNSII2 | CmFNSII3 | 0.139 352 161 | 0.427 416 010 | 0.326 034 023 |
| TD | CmDFR1 | CmDFR2 | 0.189 572 148 | 0.623 062 431 | 0.304 258 672 |
| TD | CmANR3 | CmANR4 | 0.137 094 524 | 0.393 608 899 | 0.348 301 384 |
| TD | Cm4CL4 | Cm4CL5 | 0.133 265 871 | 0.425 828 776 | 0.312 956 470 |
| TD | Cm4CL9 | Cm4CL10 | 0.103 441 680 | 0.273 134 833 | 0.378 720 204 |
| TD | CmPAL4 | CmPAL5 | 0.052 152 683 | 0.128 965 485 | 0.404 392 564 |
| TRD | CmCHS1 | CmCHS2 | 0.644 246 280 | NaN (high sequence divergence) | |
| TRD | CmCHS3 | CmCHS4 | 0.587 247 232 | 2.429 515 767 | 0.241 713 694 |
| TRD | CmLAR2 | CmLAR1 | 0.263 851 247 | 1.291 720 422 | 0.204 263 432 |
| TRD | CmF3'5'H1 | CmF3'5'H2 | 0.090 142 821 | 0.211 897 324 | 0.425 408 022 |
| TRD | CmFLS2 | CmFLS3 | 0.190 856 593 | 1.395 097 547 | 0.136 805 196 |
| TRD | CmANS2 | CmANS1 | 0.636 070 470 | 3.657 827 926 | 0.173 892 945 |
| TRD | CmFLS1 | CmFLS3 | 0.467 506 223 | 3.335 289 988 | 0.140 169 588 |
| TRD | CmANR7 | CmANR6 | 0.015 019 275 | 0.051 683 849 | 0.290 599 013 |
| TRD | Cm4CL6 | Cm4CL1 | 0.148 018 360 | 1.985 685 394 | 0.074 542 705 |
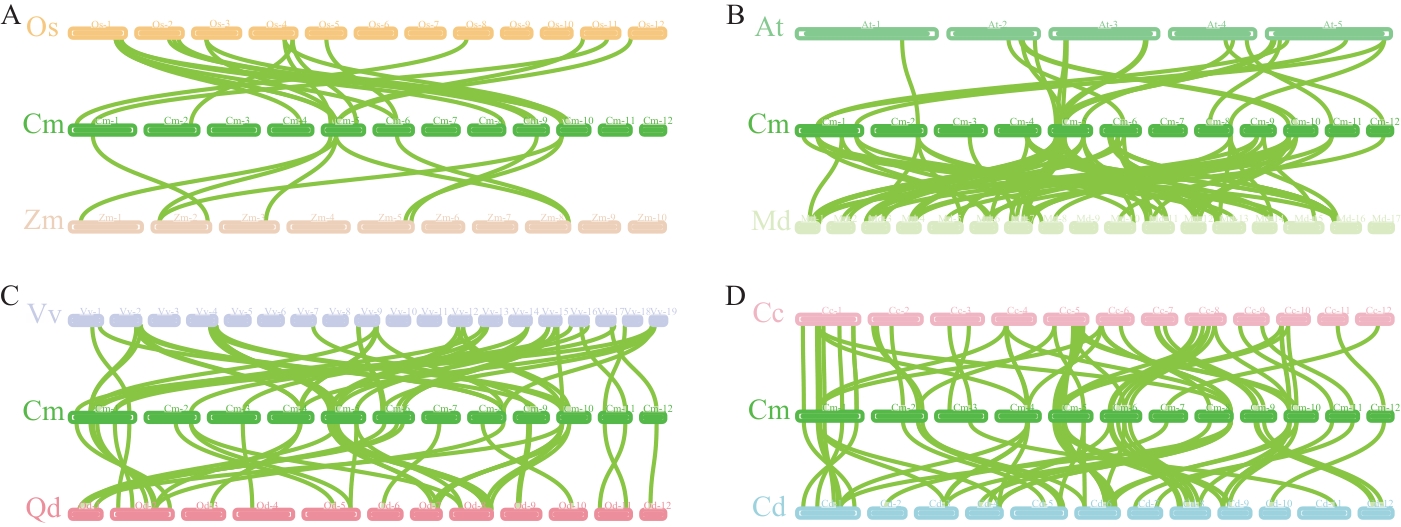
图6 板栗与物种间类黄酮合成通路基因共线性Cm:板栗;Os:水稻;Zm:玉米;At:拟南芥;Md:苹果;Vv:葡萄;Qd:槲树;Cc:日本栗;Cd:美洲栗
Fig. 6 Collinearity of flavonoid synthesis pathway genes between chestnut and speciesCm: Castanea mollissima; Os: Oryza sativa; Zm: Zea mays; At: Arabidopsis thaliana; Md: Malus domestica; Vv: Vitis vinifera; Qd: Quercus dentata; Cc: Castanea crenata; Cd: Castanea dentata

图8 板栗类黄酮合成通路基因家族成员A:中性绘图;B:ENC-plot绘图;C:PR2-plot绘图
Fig. 8 Members of the Chinese chestnut flavonoids synthesis pathway gene familyA: Neutrality curve; B: ENC-plot plot; C: PR2-plot plot

图9 板栗果实成熟过程(A)和果实顶部芽组织PCD过程(B)中类黄酮合成通路基因的表达模式Nut70、Nut82、Nut94分别代表70、82、94 d果实成熟时期; Bud20、Bud25、Bud30分别代表20、25、30 d芽组织PCD时期。下同
Fig. 9 Expression pattern of genes in flavonoid synthesis pathway in chestnut fruit ripening (A) and PCD process of fruit top bud tissue (B)Nut 70, Nut 82 and Nut 94 indicate 70, 82, and 94 d of fruit ripening period, respectively; Bud20, Bud25, and Bud30 indicate the 20, 25, and 30 d bud tissue PCD period, respectively. The same below
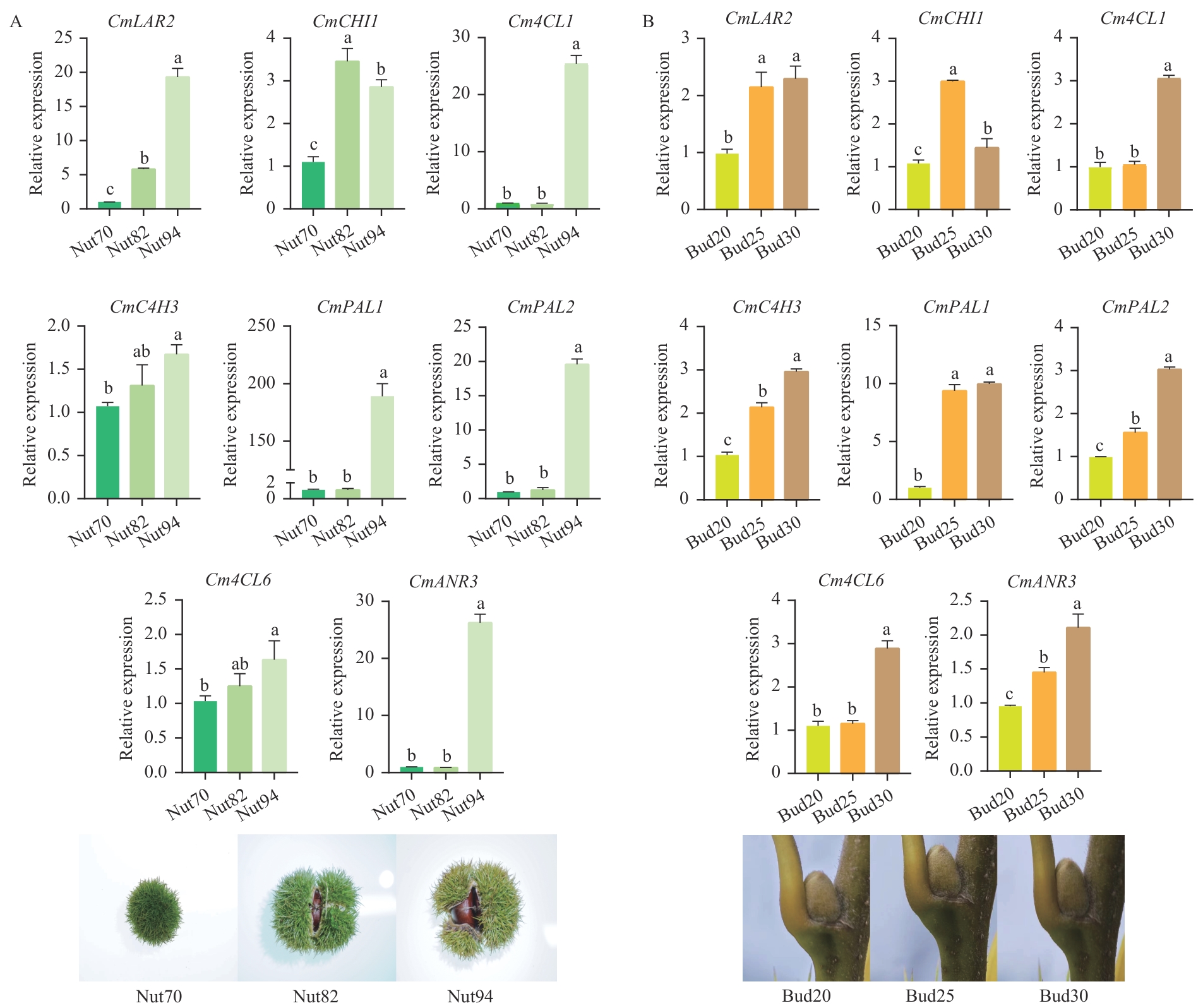
图10 板栗果实成熟过程(A)和果实顶部芽组织PCD过程(B)中类黄酮合成通路基因的相对表达量不同小写字母代表在0.05水差异显著
Fig. 10 Relative expressions of genes in flavonoid synthesis pathway in chestnut fruit ripening (A) and PCD process of fruit top bud tissue (B)Different lower letters indicate significantly difference at the 0.05 level
| 1 | Shen N, Wang TF, Gan Q, et al. Plant flavonoids: classification, distribution, biosynthesis, and antioxidant activity [J]. Food Chem, 2022, 383: 132531. |
| 2 | Li PQ, Ruan Z, Fei ZX, et al. Integrated transcriptome and metabolome analysis revealed that flavonoid biosynthesis may dominate the resistance of Zanthoxylum bungeanum against stem canker [J]. J Agric Food Chem, 2021, 69(22): 6360-6378. |
| 3 | Zhang PL, Liu J, Jia N, et al. Genome-wide identification and characterization of the bZIP gene family and their function in starch accumulation in Chinese chestnut (Castanea mollissima Blume) [J]. Front Plant Sci, 2023, 14: 1166717. |
| 4 | 樊晓芸, 郭素娟, 李艳华, 等. 板栗果实褐变度与总酚和总黄酮的相关性研究 [J]. 南京林业大学学报: 自然科学版, 2023, 47(6): 159-166. |
| Fan XY, Guo SJ, Li YH, et al. Study on correlation between browning degree of chestnut fruit and total phenols and flavonoids [J]. J Nanjing For Univ Nat Sci Ed, 2023, 47(6): 159-166. | |
| 5 | Zou J, Ge YN, Zhang Y, et al. Changes in flavor- and aroma-related fermentation metabolites and antioxidant activity of glutinous rice wine supplemented with Chinese chestnut (Castanea mollissima blume) [J]. Fermentation, 2022, 8(6): 266. |
| 6 | 黄雪薇, 雷嗣超, 涂芬, 等. 板栗壳黄酮结构分析及其对胰脂肪酶活力的抑制作用 [J]. 食品科学, 2021, 42 (21): 111-118. |
| Huang XW, Lei SC, Tu F, et al. Structural analysis and anti-pancreatic lipase activity of flavonoids from chestnut shells [J]. Food Sci, 2021, 42 (21): 111-118. | |
| 7 | Peng F, Yin HY, Du B, et al. Anti-fatigue activity of purified flavonoids prepared from chestnut (Castanea mollissima) flower [J]. J Funct Foods, 2021, 79: 104365. |
| 8 | Liu SA, Meng ZL, Zhang HY, et al. Identification and characterization of thirteen gene families involved in flavonoid biosynthesis in Ginkgo biloba [J]. Ind Crops Prod, 2022, 188: 115576. |
| 9 | 杜婷婷, 宋治华, 董碧莹, 等. 木豆类黄酮代谢通路关键基因家族的鉴定与表达分析 [J]. 农业生物技术学报, 2021, 29(12): 2289-2303. |
| Du TT, Song ZH, Dong BY, et al. Identification and expression analysis of key gene families in flavonoid metabolism pathway in pigeon pea (Cajanus cajan) [J]. J Agric Biotechnol, 2021, 29(12): 2289-2303. | |
| 10 | Wang JY, Zhang CH, Li YS. Genome-wide identification and expression profiles of 13 key structural gene families involved in the biosynthesis of rice flavonoid scaffolds [J]. Genes, 2022, 13(3): 410. |
| 11 | Deng YX, Li CL, Li HQ, et al. Identification and characterization of flavonoid biosynthetic enzyme genes in Salvia miltiorrhiza (Lamiaceae) [J]. Molecules, 2018, 23(6): 1467. |
| 12 | Han MG, Cui RF, Cui YP, et al. A flavonol synthase (FLS) gene, GhFLS1, was screened out increasing salt resistance in cotton [J]. Environ Sci Eur, 2023, 35(1): 37. |
| 13 | Wang M, Zhang Y, Zhu CY, et al. EkFLS overexpression promotes flavonoid accumulation and abiotic stress tolerance in plant [J]. Physiol Plant, 2021, 172(4): 1966-1982. |
| 14 | Hou QD, Li S, Shang CQ, et al. Genome-wide characterization of chalcone synthase genes in sweet cherry and functional characterization of CpCHS1 under drought stress [J]. Front Plant Sci, 2022, 13: 989959. |
| 15 | Zhang Y, Shu HY, Mumtaz MA, et al. Transcriptome and metabolome analysis of color changes during fruit development of pepper (Capsicum baccatum) [J]. Int J Mol Sci, 2022, 23(20): 12524. |
| 16 | Wang WQ, Pu YF, Wen H, et al. Transcriptome and weighted gene co-expression network analysis of jujube (Ziziphus jujuba Mill.) fruit reveal putative genes involved in proanthocyanin biosynthesis and regulation [J]. Food Sci Hum Wellness, 2023, 12(5): 1557-1570. |
| 17 | Li YP, Li HF, Wang SY, et al. Metabolomic and transcriptomic analyses of the flavonoid biosynthetic pathway in blueberry (Vaccinium spp.) [J]. Front Plant Sci, 2023, 14: 1082245. |
| 18 | Danon A, Delorme V, Mailhac N, et al. Plant programmed cell death: a common way to die [J]. Plant Physiol Biochem, 2000, 38(9): 647-655. |
| 19 | Petrov V, Hille J, Mueller-Roeber B, et al. ROS-mediated abiotic stress-induced programmed cell death in plants [J]. Front Plant Sci, 2015, 6: 69. |
| 20 | Wang GP, Zhang ZH, Kong DJ, et al. Programmed cell death is responsible for replaceable bud senescence in chestnut (Castanea mollissima BL.) [J]. Plant Cell Rep, 2012, 31(9): 1603-1610. |
| 21 | Guo Y, Zhang SH, Li Y, et al. A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut [J]. Open Life Sci, 2023, 18(1): 20220635. |
| 22 | Wang JP, Tian SL, Sun XL, et al. Construction of pseudomolecules for the Chinese chestnut (Castanea mollissima) genome [J]. G3, 2020, 10(10): 3565-3574. |
| 23 | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a "one for all, all for one" bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| 24 | Du LX, Lu C, Wang ZT, et al. GFAnno: integrated method for plant flavonoid biosynthesis pathway gene annotation [J]. Beverage Plant Res, 2024, 4(1). |
| 25 | Rozewicki J, Li SL, Amada KM, et al. MAFFT-DASH: integrated protein sequence and structural alignment [J]. Nucleic Acids Res, 2019, 47(W1): W5-W10. |
| 26 | Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments [J]. PLoS One, 2010, 5(3): e9490. |
| 27 | Xie JM, Chen YR, Cai GJ, et al. Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees [J]. Nucleic Acids Res, 2023, 51(W1): W587-W592. |
| 28 | Wang YP, Tang HB, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity [J]. Nucleic Acids Res, 2012, 40(7): e49. |
| 29 | Qiao X, Li QH, Yin H, et al. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants [J]. Genome Biol, 2019, 20(1): 38. |
| 30 | Gao Y, Lu Y, Song Y, et al. Analysis of codon usage bias of WRKY transcription factors in Helianthus annuus [J]. BMC Genom Data, 2022, 23(1): 46. |
| 31 | Zhang Y, Shen ZN, Meng XR, et al. Codon usage patterns across seven Rosales species [J]. BMC Plant Biol, 2022, 22(1): 65. |
| 32 | Li Q, Luo YY, Sha AJ, et al. Analysis of synonymous codon usage patterns in mitochondrial genomes of nine Amanita species [J]. Front Microbiol, 2023, 14: 1134228. |
| 33 | Yu LY, Fei C, Wang DS, et al. Genome-wide identification, evolution and expression profiles analysis of bHLH gene family in Castanea mollissima [J]. Front Genet, 2023, 14: 1193953. |
| 34 | 赵奇, 茹京娜, 李宜统, 等. 小麦Lhc基因家族鉴定与表达模式分析 [J]. 植物遗传资源学报, 2022, 23(6): 1766-1781. |
| Zhao Q, Ru JN, Li YT, et al. Identification and expression pattern analysis of Lhc gene family members in wheat [J]. J Plant Genet Resour, 2022, 23(6): 1766-1781. | |
| 35 | Liu H, Lyu HM, Zhu KK, et al. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families [J]. Plant J, 2021, 105(4): 1072-1082. |
| 36 | Zhang LC, Song BB, Li B, et al. Genome-wide identification and expression analysis of fifteen gene families involved in anthocyanin synthesis in pear [J]. Horticulturae, 2024, 10(4): 335. |
| 37 | Yang WL, Li N, Fan YX, et al. Transcriptome analysis reveals abscisic acid enhancing drought resistance by regulating genes related to flavonoid metabolism in pigeon pea [J]. Environ Exp Bot, 2021, 191: 104627. |
| 38 | Eom SH, Ahn MA, Kim E, et al. Plant response to cold stress: cold stress changes antioxidant metabolism in heading type kimchi cabbage (Brassica rapa L. ssp. pekinensis) [J]. Antioxidants, 2022, 11(4): 700. |
| 39 | Wang CG, Zhang MY, Zhou JJ, et al. Transcriptome analysis and differential gene expression profiling of Wucai (Brassica campestris L.) in response to cold stress [J]. BMC Genomics, 2022, 23(1): 137. |
| 40 | Qu JJ, Liu LL, Guo ZX, et al. The ubiquitous position effect, synergistic effect of recent generated tandem duplicated genes in grapevine, and their co-response and overactivity to biotic stress [J]. Fruit Res, 2023, 3(1). |
| 41 | Fang C, Yang MY, Tang YC, et al. Dynamics of cis-regulatory sequences and transcriptional divergence of duplicated genes in soybean [J]. Proc Natl Acad Sci U S A, 2023, 120(44): e2303836120. |
| 42 | Xu YH, Zhang KJ, Zhang ZL, et al. A chromosome-level genome assembly for Dracaena cochinchinensis reveals the molecular basis of its longevity and formation of dragon's blood [J]. Plant Commun, 2022, 3(6): 100456. |
| [1] | 颜伟, 陈慧婷, 叶青, 刘广超, 刘新, 侯丽霞. 葡萄HCT基因家族鉴定及其对低温胁迫的响应[J]. 生物技术通报, 2025, 41(2): 175-186. |
| [2] | 匡健华, 程志鹏, 赵永晶, 杨洁, 陈润乔, 陈龙清, 胡慧贞. 激素和非生物胁迫下荷花GH3基因家族的表达分析[J]. 生物技术通报, 2025, 41(2): 221-233. |
| [3] | 杨涌, 袁国梅, 康肖肖, 刘亚明, 王东升, 张海娥. 板栗SWEET基因家族成员的鉴定及表达分析[J]. 生物技术通报, 2025, 41(2): 257-269. |
| [4] | 李禹欣, 李苗, 杜晓芬, 韩康妮, 连世超, 王军. 谷子SiSAP基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(1): 143-156. |
| [5] | 孔青洋, 张晓龙, 李娜, 张晨洁, 张雪云, 于超, 张启翔, 罗乐. 单叶蔷薇GRAS转录因子家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 210-220. |
| [6] | 申鹏, 高雅彬, 丁红. 马铃薯SAT基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(9): 64-73. |
| [7] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| [8] | 吴慧琴, 王延宏, 刘涵, 司政, 刘雪晴, 王静, 阳宜, 成妍. 辣椒UGT基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 198-211. |
| [9] | 谭博文, 张懿, 张鹏, 王振宇, 马秋香. 木薯镁离子转运蛋白家族基因的鉴定及生物信息学分析[J]. 生物技术通报, 2024, 40(9): 20-32. |
| [10] | 王茜, 周家燕, 王倩, 邓玉萍, 张敏慧, 陈静, 杨军, 邹建. 向日葵YABBY基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(8): 199-211. |
| [11] | 李勇慧, 鲍星星, 段一珂, 赵运霞, 于相丽, 陈尧, 张延召. 灵宝杜鹃bZIP家族全基因组鉴定及表达特征分析[J]. 生物技术通报, 2024, 40(8): 186-198. |
| [12] | 周麟, 黄顺满, 苏文坤, 姚响, 屈燕. 滇山茶bHLH基因家族鉴定及花色形成相关基因筛选[J]. 生物技术通报, 2024, 40(8): 142-151. |
| [13] | 张明亚, 庞胜群, 刘玉东, 苏永峰, 牛博文, 韩琼琼. 番茄FAD基因家族的鉴定与表达分析[J]. 生物技术通报, 2024, 40(7): 150-162. |
| [14] | 吴泽航, 杨中义, 鄢毅铖, 贾永红, 吴月燕, 谢晓鸿. 比利时杜鹃花类黄酮3'-羟化酶(F3'H)基因克隆及功能分析[J]. 生物技术通报, 2024, 40(6): 251-259. |
| [15] | 胡永波, 雷雨田, 杨永森, 陈馨, 林黄昉, 林碧英, 刘爽, 毕格, 申宝营. 黄瓜和南瓜Bcl-2相关抗凋亡家族全基因组鉴定与表达模式分析[J]. 生物技术通报, 2024, 40(6): 219-237. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||