








生物技术通报 ›› 2024, Vol. 40 ›› Issue (9): 20-32.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0521
• 薯类作物生物技术专题(专题主编:徐建飞,尚轶) • 上一篇 下一篇
谭博文1,2( ), 张懿2,3, 张鹏2,3, 王振宇4(
), 张懿2,3, 张鹏2,3, 王振宇4( ), 马秋香2(
), 马秋香2( )
)
收稿日期:2024-05-30
出版日期:2024-09-26
发布日期:2024-10-12
通讯作者:
王振宇,男,教授,研究方向:植物分子育种;E-mail: wangzy80@126.com;作者简介:谭博文,男,硕士研究生,研究方向:薯类分子育种;E-mail: 1050565123@qq.com
基金资助:
TAN Bo-wen1,2( ), ZHANG Yi2,3, ZHANG Peng2,3, WANG Zhen-yu4(
), ZHANG Yi2,3, ZHANG Peng2,3, WANG Zhen-yu4( ), MA Qiu-xiang2(
), MA Qiu-xiang2( )
)
Received:2024-05-30
Published:2024-09-26
Online:2024-10-12
摘要:
【目的】镁离子转运蛋白(magnesium transporters, MGT)是重要的镁离子运输体,通过分析木薯MeMGT家族基因的基本信息及所编码蛋白质的物理化学特性,为进一步研究其功能提供指导。【方法】通过同源序列比对,从木薯基因组数据库中筛选出13个MeMGTs基因,分别将其命名为MeMGT2;1-MeMGT10;2。运用生物信息学方法对这些基因进行染色体定位分析、保守序列功能预测和系统发育树分析,并对所编码的蛋白质理化性质和结构功能进行预测。【结果】染色体定位分析显示13个MeMGTs基因分布在8条染色体上,基因的外显子数量在4-13个之间。组织表达模式分析表明MeMGTs基因在不同发育阶段表达水平不同,其中MeMGT4;2主要在须根和储藏根中表达,推测该基因可能主要在根的发育过程中发挥作用;MeMGT7主要在木薯叶片、中脉及茎中表达,推测其可能主要在镁离子的长距离运输过程中发挥作用。顺式作用元件预测结果显示MeMGTs基因的启动子序列中含有大量光响应和激素响应元件,暗示其受多种因子调控。分析木薯与部分模式植物MGT基因家族的系统发育树,发现这些蛋白被分为5类。蛋白序列特征分析显示MeMGTs蛋白均为亲水性蛋白,具有2个跨膜结构和保守的镁转运蛋白三肽基序Gly-Met-Asn(GMN),且不含信号肽。蛋白磷酸化分析表明MeMGTs基因家族具有丝氨酸、苏氨酸和酪氨酸磷酸化位点,该基因家族的蛋白可能通过这些位点发挥作用。【结论】MeMGTs基因表达受光、激素、干旱和低温响应等多种元件调控,可能在木薯的生长发育和逆境胁迫中发挥用。
谭博文, 张懿, 张鹏, 王振宇, 马秋香. 木薯镁离子转运蛋白家族基因的鉴定及生物信息学分析[J]. 生物技术通报, 2024, 40(9): 20-32.
TAN Bo-wen, ZHANG Yi, ZHANG Peng, WANG Zhen-yu, MA Qiu-xiang. Identification and Bioinformatics Analysis of Gene in the Magnesium Transporter Family in Cassava[J]. Biotechnology Bulletin, 2024, 40(9): 20-32.
| 基因Gene | CAAT-box | TATA-box | 光响应元件Light responsive elements | 激素响应元件Hormone responsive elements | 干旱胁迫响应元件Drought stress responsive elements | 低温响应元件Low temperature responsive elements | 其他Others |
|---|---|---|---|---|---|---|---|
| MeMGT2;1 | 13 | 104 | 23 | 2 | 0 | 1 | 8 |
| MeMGT2;2 | 14 | 48 | 15 | 4 | 1 | 3 | 15 |
| MeMGT3;1 | 16 | 41 | 6 | 3 | 0 | 0 | 27 |
| MeMGT3;2 | 17 | 78 | 14 | 5 | 1 | 0 | 12 |
| MeMGT4;1 | 20 | 92 | 9 | 3 | 2 | 0 | 14 |
| MeMGT4;2 | 6 | 168 | 14 | 3 | 3 | 1 | 8 |
| MeMGT6 | 9 | 111 | 17 | 3 | 0 | 1 | 13 |
| MeMGT7 | 7 | 103 | 15 | 6 | 0 | 0 | 13 |
| MeMGT9;1 | 10 | 48 | 12 | 5 | 1 | 1 | 20 |
| MeMGT9;2 | 17 | 70 | 18 | 5 | 0 | 2 | 9 |
| MeMGT9;3 | 22 | 61 | 20 | 4 | 0 | 0 | 17 |
| MeMGT10;1 | 16 | 71 | 4 | 9 | 1 | 0 | 27 |
| MeMGT10;2 | 13 | 85 | 20 | 6 | 2 | 0 | 20 |
表1 木薯MeMGTs基因家族顺式作用元件分析
Table 1 Analysis of cis-acting elements of MeMGTs gene family in cassava
| 基因Gene | CAAT-box | TATA-box | 光响应元件Light responsive elements | 激素响应元件Hormone responsive elements | 干旱胁迫响应元件Drought stress responsive elements | 低温响应元件Low temperature responsive elements | 其他Others |
|---|---|---|---|---|---|---|---|
| MeMGT2;1 | 13 | 104 | 23 | 2 | 0 | 1 | 8 |
| MeMGT2;2 | 14 | 48 | 15 | 4 | 1 | 3 | 15 |
| MeMGT3;1 | 16 | 41 | 6 | 3 | 0 | 0 | 27 |
| MeMGT3;2 | 17 | 78 | 14 | 5 | 1 | 0 | 12 |
| MeMGT4;1 | 20 | 92 | 9 | 3 | 2 | 0 | 14 |
| MeMGT4;2 | 6 | 168 | 14 | 3 | 3 | 1 | 8 |
| MeMGT6 | 9 | 111 | 17 | 3 | 0 | 1 | 13 |
| MeMGT7 | 7 | 103 | 15 | 6 | 0 | 0 | 13 |
| MeMGT9;1 | 10 | 48 | 12 | 5 | 1 | 1 | 20 |
| MeMGT9;2 | 17 | 70 | 18 | 5 | 0 | 2 | 9 |
| MeMGT9;3 | 22 | 61 | 20 | 4 | 0 | 0 | 17 |
| MeMGT10;1 | 16 | 71 | 4 | 9 | 1 | 0 | 27 |
| MeMGT10;2 | 13 | 85 | 20 | 6 | 2 | 0 | 20 |
| 基因Gene | 氨基酸数Amino-acid number | 蛋白质分子量Molecular weight/Da | 等电点Isoelectric point | 不稳定系数Instability index | 脂肪指数Aliphaic index | 亚细胞定位Subcellular localization |
|---|---|---|---|---|---|---|
| MeMGT2;1 | 441 | 5 116.63 | 5.24 | 61.16 | 99.71 | Plasma membrane |
| MeMGT2;2 | 443 | 50 084.46 | 5.17 | 60.79 | 99.68 | Plasma membrane |
| MeMGT3;1 | 419 | 46 929.10 | 5.31 | 41.71 | 105.89 | Plasma membrane |
| MeMGT3;2 | 420 | 47 276.18 | 5.15 | 39.36 | 100.98 | Plasma membrane |
| MeMGT4;1 | 495 | 55 072.03 | 4.85 | 41.36 | 91.23 | Cytoplasmic |
| MeMGT4;2 | 496 | 55 057.13 | 4.94 | 42.90 | 89.46 | Cytoplasmic |
| MeMGT6 | 449 | 50 165.42 | 5.59 | 48.10 | 100.56 | Chloroplast |
| MeMGT7 | 391 | 44 564.62 | 4.87 | 34.30 | 102.23 | Golgi apparatus |
| MeMGT9;1 | 400 | 44 715.05 | 5.07 | 40.07 | 99.27 | Plasma membrane |
| MeMGT9;2 | 402 | 44 505.71 | 4.92 | 41.99 | 100.00 | Chloroplast |
| MeMGT9;3 | 423 | 47 245.86 | 5.06 | 51.35 | 97.09 | Cytoplasmic |
| MeMGT10;1 | 444 | 49 771.50 | 5.82 | 54.37 | 106.01 | Plasma membrane |
| MeMGT10;2 | 447 | 50 089.05 | 5.75 | 54.74 | 107.25 | Plasma membrane |
表2 木薯MeMGTs蛋白物理化学性质
Table 2 Physicochemical properties of MeMGTs proteins in cassava
| 基因Gene | 氨基酸数Amino-acid number | 蛋白质分子量Molecular weight/Da | 等电点Isoelectric point | 不稳定系数Instability index | 脂肪指数Aliphaic index | 亚细胞定位Subcellular localization |
|---|---|---|---|---|---|---|
| MeMGT2;1 | 441 | 5 116.63 | 5.24 | 61.16 | 99.71 | Plasma membrane |
| MeMGT2;2 | 443 | 50 084.46 | 5.17 | 60.79 | 99.68 | Plasma membrane |
| MeMGT3;1 | 419 | 46 929.10 | 5.31 | 41.71 | 105.89 | Plasma membrane |
| MeMGT3;2 | 420 | 47 276.18 | 5.15 | 39.36 | 100.98 | Plasma membrane |
| MeMGT4;1 | 495 | 55 072.03 | 4.85 | 41.36 | 91.23 | Cytoplasmic |
| MeMGT4;2 | 496 | 55 057.13 | 4.94 | 42.90 | 89.46 | Cytoplasmic |
| MeMGT6 | 449 | 50 165.42 | 5.59 | 48.10 | 100.56 | Chloroplast |
| MeMGT7 | 391 | 44 564.62 | 4.87 | 34.30 | 102.23 | Golgi apparatus |
| MeMGT9;1 | 400 | 44 715.05 | 5.07 | 40.07 | 99.27 | Plasma membrane |
| MeMGT9;2 | 402 | 44 505.71 | 4.92 | 41.99 | 100.00 | Chloroplast |
| MeMGT9;3 | 423 | 47 245.86 | 5.06 | 51.35 | 97.09 | Cytoplasmic |
| MeMGT10;1 | 444 | 49 771.50 | 5.82 | 54.37 | 106.01 | Plasma membrane |
| MeMGT10;2 | 447 | 50 089.05 | 5.75 | 54.74 | 107.25 | Plasma membrane |
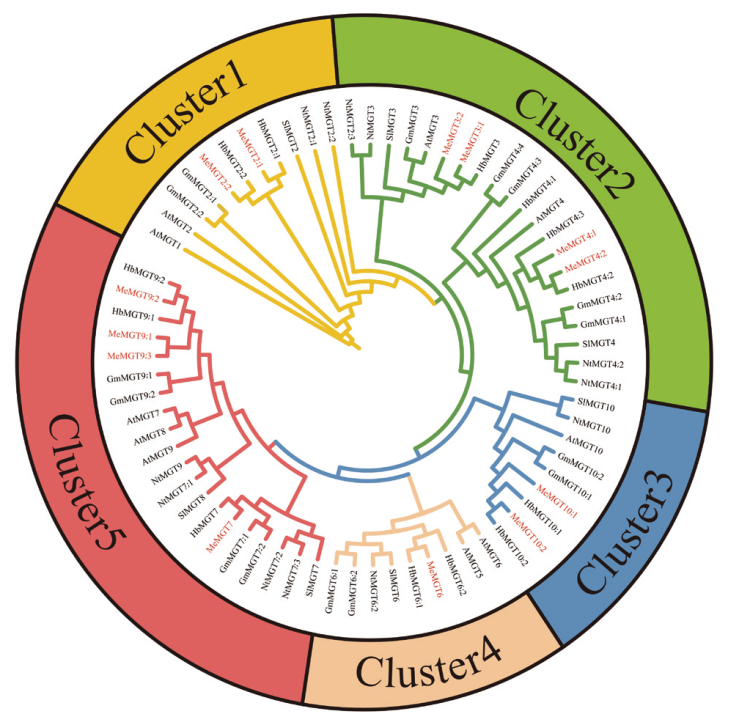
图4 木薯、橡胶树、拟南芥、烟草、大豆、番茄MGT基因家族成员进化树
Fig. 4 Evolutionary tree of MGT gene family members of Manihot esculenta crantz, Hevea brasiliensis, Arabi-dopsis thaliana, Nicotiana tabacum, Glycine max, and Solanum lycopersicum

图7 木薯MeMGTs蛋白保守区段分析 图中黑框标记为镁转运蛋白保守区域三肽基序Gly-Met-Asn
Fig. 7 Conservative analysis of MeMGTs proteins in cassava The black box in the figure is marked with the tripeptide motif Gly-Met-Asn in the conserved region of the magnesium transporter
| 基因Gene | 基因ID Gene ID | MeMGTs基因家族蛋白质二级结构预测Prediction of secondary structure of MeMGTs gene family proteins/% | ||||
|---|---|---|---|---|---|---|
| α-螺旋α-helix | 延伸链Extended strand | β-转角β-turn | 无规则卷曲Random coil | |||
| MeMGT2;1 | Manes.11G068600 | 53.74 | 6.80 | 2.04 | 37.41 | |
| MeMGT2;2 | Manes.04G101500 | 51.02 | 7.45 | 2.93 | 38.60 | |
| MeMGT3;1 | Manes.05G110400 | 54.89 | 7.16 | 2.63 | 35.32 | |
| MeMGT3;2 | Manes.01G027400 | 54.05 | 7.62 | 2.38 | 35.95 | |
| MeMGT4;1 | Manes.07G053800 | 52.93 | 5.45 | 2.22 | 39.39 | |
| MeMGT4;2 | Manes.10G084800 | 55.44 | 4.64 | 1.21 | 38.71 | |
| MeMGT6 | Manes.07G020300 | 50.78 | 6.68 | 2.23 | 40.31 | |
| MeMGT7 | Manes.05G085100 | 57.80 | 8.44 | 3.32 | 30.43 | |
| MeMGT9;1 | Manes.03G169600 | 60.00 | 7.50 | 2.50 | 30.00 | |
| MeMGT9;2 | Manes.15G036500 | 59.20 | 6.97 | 3.23 | 30.60 | |
| MeMGT9;3 | Manes.15G036400 | 56.50 | 9.22 | 4.26 | 30.02 | |
| MeMGT10;1 | Manes.11G155200 | 47.52 | 8.33 | 2.93 | 41.22 | |
| MeMGT10;2 | Manes.04G011000 | 51.01 | 7.38 | 2.46 | 39.15 | |
表3 木薯MeMGTs蛋白氨基酸序列的二级结构元件组成
Table 3 Composition of predicted secondary structure among MeMGTs proteins in cassava
| 基因Gene | 基因ID Gene ID | MeMGTs基因家族蛋白质二级结构预测Prediction of secondary structure of MeMGTs gene family proteins/% | ||||
|---|---|---|---|---|---|---|
| α-螺旋α-helix | 延伸链Extended strand | β-转角β-turn | 无规则卷曲Random coil | |||
| MeMGT2;1 | Manes.11G068600 | 53.74 | 6.80 | 2.04 | 37.41 | |
| MeMGT2;2 | Manes.04G101500 | 51.02 | 7.45 | 2.93 | 38.60 | |
| MeMGT3;1 | Manes.05G110400 | 54.89 | 7.16 | 2.63 | 35.32 | |
| MeMGT3;2 | Manes.01G027400 | 54.05 | 7.62 | 2.38 | 35.95 | |
| MeMGT4;1 | Manes.07G053800 | 52.93 | 5.45 | 2.22 | 39.39 | |
| MeMGT4;2 | Manes.10G084800 | 55.44 | 4.64 | 1.21 | 38.71 | |
| MeMGT6 | Manes.07G020300 | 50.78 | 6.68 | 2.23 | 40.31 | |
| MeMGT7 | Manes.05G085100 | 57.80 | 8.44 | 3.32 | 30.43 | |
| MeMGT9;1 | Manes.03G169600 | 60.00 | 7.50 | 2.50 | 30.00 | |
| MeMGT9;2 | Manes.15G036500 | 59.20 | 6.97 | 3.23 | 30.60 | |
| MeMGT9;3 | Manes.15G036400 | 56.50 | 9.22 | 4.26 | 30.02 | |
| MeMGT10;1 | Manes.11G155200 | 47.52 | 8.33 | 2.93 | 41.22 | |
| MeMGT10;2 | Manes.04G011000 | 51.01 | 7.38 | 2.46 | 39.15 | |
| 基因Gene | 基因ID Gene ID | MeMGTs基因家族蛋白质磷酸化位点数量Number of phosphorylation sites of MeMGTs gene family proteins | ||
|---|---|---|---|---|
| 丝氨酸Ser | 苏氨酸Thr | 酪氨酸Tyr | ||
| MeMGT2;1 | Manes.11G068600 | 33 | 9 | 4 |
| MeMGT2;2 | Manes.04G101500 | 33 | 11 | 3 |
| MeMGT3;1 | Manes.05G110400 | 18 | 15 | 4 |
| MeMGT3;2 | Manes.01G027400 | 22 | 14 | 4 |
| MeMGT4;1 | Manes.07G053800 | 16 | 20 | 3 |
| MeMGT4;2 | Manes.10G084800 | 25 | 25 | 3 |
| MeMGT6 | Manes.07G020300 | 22 | 13 | 3 |
| MeMGT7 | Manes.05G085100 | 22 | 17 | 6 |
| MeMGT9;1 | Manes.03G169600 | 19 | 13 | 3 |
| MeMGT9;2 | Manes.15G036500 | 19 | 12 | 5 |
| MeMGT9;3 | Manes.15G036400 | 22 | 12 | 2 |
| MeMGT10;1 | Manes.11G155200 | 27 | 10 | 3 |
| MeMGT10;2 | Manes.04G011000 | 31 | 7 | 4 |
表4 木薯MeMGTs蛋白磷酸化位点数量
Table 4 Number of phosphorylation sites of MeMGTs proteins in cassava
| 基因Gene | 基因ID Gene ID | MeMGTs基因家族蛋白质磷酸化位点数量Number of phosphorylation sites of MeMGTs gene family proteins | ||
|---|---|---|---|---|
| 丝氨酸Ser | 苏氨酸Thr | 酪氨酸Tyr | ||
| MeMGT2;1 | Manes.11G068600 | 33 | 9 | 4 |
| MeMGT2;2 | Manes.04G101500 | 33 | 11 | 3 |
| MeMGT3;1 | Manes.05G110400 | 18 | 15 | 4 |
| MeMGT3;2 | Manes.01G027400 | 22 | 14 | 4 |
| MeMGT4;1 | Manes.07G053800 | 16 | 20 | 3 |
| MeMGT4;2 | Manes.10G084800 | 25 | 25 | 3 |
| MeMGT6 | Manes.07G020300 | 22 | 13 | 3 |
| MeMGT7 | Manes.05G085100 | 22 | 17 | 6 |
| MeMGT9;1 | Manes.03G169600 | 19 | 13 | 3 |
| MeMGT9;2 | Manes.15G036500 | 19 | 12 | 5 |
| MeMGT9;3 | Manes.15G036400 | 22 | 12 | 2 |
| MeMGT10;1 | Manes.11G155200 | 27 | 10 | 3 |
| MeMGT10;2 | Manes.04G011000 | 31 | 7 | 4 |
| [1] | Wilkinson SR, Welch R, Mayland HF, et al. Magnesium in plants: uptake, distribution, function, and utilization by man and animals[J]. Met Ions Biol Syst, 1990, 26: 33-56. |
| [2] |
Chen ZC, Peng WT, Li J, et al. Functional dissection and transport mechanism of magnesium in plants[J]. Semin Cell Dev Biol, 2018, 74: 142-152.
doi: S1084-9521(17)30257-4 pmid: 28822768 |
| [3] |
Hermans C, Conn SJ, Chen JG, et al. An update on magnesium homeostasis mechanisms in plants[J]. Metallomics, 2013, 5(9): 1170-1183.
doi: 10.1039/c3mt20223b pmid: 23420558 |
| [4] |
Kuhn AJ, Schröder WH, Bauch J. The kinetics of calcium and magnesium entry into mycorrhizal spruce roots[J]. Planta, 2000, 210(3): 488-496.
pmid: 10750907 |
| [5] | Chaudhry AH, Nayab S, Hussain SB, et al. Current understandings on magnesium deficiency and future outlooks for sustainable agriculture[J]. Int J Mol Sci, 2021, 22(4): 1819. |
| [6] |
Zhang BG, Cakmak I, Feng JC, et al. Magnesium deficiency reduced the yield and seed germination in wax gourd by affecting the carbohydrate translocation[J]. Front Plant Sci, 2020, 11: 797.
doi: 10.3389/fpls.2020.00797 pmid: 32595681 |
| [7] | Ahmed N, Zhang BG, Bozdar B, et al. The power of magnesium: unlocking the potential for increased yield, quality, and stress tolerance of horticultural crops[J]. Front Plant Sci, 2023, 14: 1285512. |
| [8] | Mengutay M, Ceylan Y, Kutman UB, et al. Adequate magnesium nutrition mitigates adverse effects of heat stress on maize and wheat[J]. Plant Soil, 2013, 368(1): 57-72. |
| [9] |
Li L, Tutone AF, Drummond RS, et al. A novel family of magnesium transport genes in Arabidopsis[J]. Plant Cell, 2001, 13(12): 2761-2775.
doi: 10.1105/tpc.010352 pmid: 11752386 |
| [10] |
Saito T, Kobayashi NI, Tanoi K, et al. Expression and functional analysis of the CorA-MRS2-ALR-type magnesium transporter family in rice[J]. Plant Cell Physiol, 2013, 54(10): 1673-1683.
doi: 10.1093/pcp/pct112 pmid: 23926064 |
| [11] |
Li HY, Du HM, Huang KF, et al. Identification, and functional and expression analyses of the CorA/MRS2/MGT-type magnesium transporter family in maize[J]. Plant Cell Physiol, 2016, 57(6): 1153-1168.
doi: 10.1093/pcp/pcw064 pmid: 27084594 |
| [12] | Li HY, Liu C, Zhou LN, et al. Molecular and functional characterization of the magnesium transporter gene ZmMGT12 in maize[J]. Gene, 2018, 665: 167-173. |
| [13] | Mao DD, Chen J, Tian LF, et al. Arabidopsis transporter MGT6 mediates magnesium uptake and is required for growth under magnesium limitation[J]. Plant Cell, 2014, 26(5): 2234-2248. |
| [14] | Mao DD, Tian LF, Li LG, et al. AtMGT7: an Arabidopsis gene encoding a low-affinity magnesium transporter[J]. J Integr Plant Biol, 2008, 50(12): 1530-1538. |
| [15] |
Deng W, Luo KM, Li DM, et al. Overexpression of an Arabidopsis magnesium transport gene, AtMGT1, in Nicotiana benthamiana confers Al tolerance[J]. J Exp Bot, 2006, 57(15): 4235-4243.
doi: 10.1093/jxb/erl201 pmid: 17101715 |
| [16] | Chen J, Li LG, Liu ZH, et al. Magnesium transporter AtMGT9 is essential for pollen development in Arabidopsis[J]. Cell Res, 2009, 19(7): 887-898. |
| [17] | Xu XF, Wang B, Lou Y, et al. Magnesium Transporter 5 plays an important role in Mg transport for male gametophyte development in Arabidopsis[J]. Plant J, 2015, 84(5): 925-936. |
| [18] |
Chen ZC, Yamaji N, Horie T, et al. A magnesium transporter OsMGT1 plays a critical role in salt tolerance in rice[J]. Plant Physiol, 2017, 174(3): 1837-1849.
doi: 10.1104/pp.17.00532 pmid: 28487477 |
| [19] | Tang L, Xiao LD, Chen EX, et al. Magnesium transporter CsMGT10 of tea plants plays a key role in chlorosis leaf vein greening[J]. Plant Physiol Biochem, 2023, 201: 107842. |
| [20] | Ma QX, Feng YC, Luo S, et al. The aquaporin MePIP2;7 improves MeMGT9-mediated Mg2+ acquisition in cassava[J]. J Integr Plant Biol, 2023, 65(10): 2349-2367. |
| [21] | 张鹏. 能源木薯种质资源面临的问题与解决策略[J]. 生物产业技术, 2008(5): 25-30. |
| Zhang P. Problems faced by energy cassava germplasm resources and solutions[J]. Biotechnology & Business, 2008(5): 25-30. | |
| [22] |
El-Sharkawy MA. Cassava biology and physiology[J]. Plant Mol Biol, 2004, 56(4): 481-501.
doi: 10.1007/s11103-005-2270-7 pmid: 15669146 |
| [23] |
Lavigne R, Seto D, Mahadevan P, et al. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools[J]. Res Microbiol, 2008, 159(5): 406-414.
doi: 10.1016/j.resmic.2008.03.005 pmid: 18555669 |
| [24] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [25] |
Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics[J]. Genome Res, 2009, 19(9): 1639-1645.
doi: 10.1101/gr.092759.109 pmid: 19541911 |
| [26] |
Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1): 325-327.
doi: 10.1093/nar/30.1.325 pmid: 11752327 |
| [27] |
Pertea M, Kim D, Pertea GM, et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown[J]. Nat Protoc, 2016, 11(9): 1650-1667.
doi: 10.1038/nprot.2016.095 pmid: 27560171 |
| [28] | Kolde R. Pheatmap: Pretty Heatmaps. R package version 1.0.12. 2019. |
| [29] | Gasteiger E, Hoogland C, Gattiker A, et al. Protein identification and analysis tools on the ExPASy server[M]//The Proteomics Protocols Handbook. Totowa, NJ: Humana Press, 2005: 571-607. |
| [30] |
Katoh K, Misawa K, Kuma KI, et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform[J]. Nucleic Acids Res, 2002, 30(14): 3059-3066.
doi: 10.1093/nar/gkf436 pmid: 12136088 |
| [31] |
Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era[J]. Mol Biol Evol, 2020, 37(5): 1530-1534.
doi: 10.1093/molbev/msaa015 pmid: 32011700 |
| [32] | Letunic I, Bork P. Interactive Tree Of Life(iTOL)v5: an online tool for phylogenetic tree display and annotation[J]. Nucleic Acids Res, 2021, 49(W1): W293-W296. |
| [33] |
Gouet P, Courcelle E, Stuart DI, et al. ESPript: analysis of multiple sequence alignments in PostScript[J]. Bioinformatics, 1999, 15(4): 305-308.
doi: 10.1093/bioinformatics/15.4.305 pmid: 10320398 |
| [34] |
Möller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions[J]. Bioinformatics, 2001, 17(7): 646-653.
doi: 10.1093/bioinformatics/17.7.646 pmid: 11448883 |
| [35] |
Krogh A, Larsson B, von Heijne G, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes[J]. J Mol Biol, 2001, 305(3): 567-580.
doi: 10.1006/jmbi.2000.4315 pmid: 11152613 |
| [36] | Bailey TL, Johnson J, Grant CE, et al. The MEME suite[J]. Nucleic Acids Res, 2015, 43(w1): W39-W49. |
| [37] | Marchler-Bauer A, Derbyshire MK, Gonzales NR, et al. CDD: NCBI's conserved domain database[J]. Nucleic Acids Res, 2015, 43(D1): D222-D226. |
| [38] | Geourjon C, Deléage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments[J]. Comput Appl Biosci, 1995, 11(6): 681-684. |
| [39] | Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes[J]. Nucleic Acids Res, 2018, 46(W1): W296-W303. |
| [40] | Bienert S, Waterhouse A, de Beer TAP, et al. The SWISS-MODEL Repository-new features and functionality[J]. Nucleic Acids Res, 2017, 45(D1): D313-D319. |
| [41] | Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective[J]. Electrophoresis, 2009, 30(Suppl 1): S162-S173. |
| [42] | Studer G, Rempfer C, Waterhouse AM, et al. QMEANDisCo—distance constraints applied on model quality estimation[J]. Bioinformatics, 2020, 36(6): 1765-1771. |
| [43] |
Bertoni M, Kiefer F, Biasini M, et al. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology[J]. Sci Rep, 2017, 7(1): 10480.
doi: 10.1038/s41598-017-09654-8 pmid: 28874689 |
| [44] |
Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites[J]. J Mol Biol, 1999, 294(5): 1351-1362.
doi: 10.1006/jmbi.1999.3310 pmid: 10600390 |
| [45] | Li HY, Wang N, Ding JZ, et al. The maize CorA/MRS2/MGT-type Mg transporter, ZmMGT10, responses to magnesium deficiency and confers low magnesium tolerance in transgenic Arabidopsis[J]. Plant Mol Biol, 2017, 95(3): 269-278. |
| [46] | Tang YH, Yang XY, Li H, et al. Uncovering the role of wheat magnesium transporter family genes in abiotic responses[J]. Front Plant Sci, 2023, 14: 1078299. |
| [47] | Reza MH, Shah H, Manjrekar J, et al. Magnesium uptake by CorA transporters is essential for growth, development and infection in the rice blast fungus Magnaporthe oryzae[J]. PLoS One, 2016, 11(7): e0159244. |
| [48] | Ishfaq M, Zhong YT, Wang YQ, et al. Magnesium limitation leads to transcriptional down-tuning of auxin synthesis, transport, and signaling in the tomato root[J]. Front Plant Sci, 2021, 12: 802399. |
| [49] | Min XJ, Powell B, Braessler J, et al. Genome-wide cataloging and analysis of alternatively spliced genes in cereal crops[J]. BMC Genomics, 2015, 16(1): 721. |
| [50] | Yang XY, Kobayashi NI, Hayashi Y, et al. Mutagenesis analysis of GMN motif in Arabidopsis thaliana Mg2+ transporter MRS2-1[J]. Biosci Biotechnol Biochem, 2022, 86(7): 870-874. |
| [51] | Schmitz J, Tierbach A, Lenz H, et al. Membrane protein interactions between different Arabidopsis thaliana MRS2-type magnesium transporters are highly permissive[J]. Biochim Biophys Acta, 2013, 1828(9): 2032-2040. |
| [52] | Ishijima S, Shiomi R, Sagami I. Functional analysis of whether the glycine residue of the GMN motif of the Arabidopsis MRS2/MGT/CorA-type Mg2+ channel protein AtMRS2-11 is critical for Mg2+ transport activity[J]. Arch Biochem Biophys, 2021, 697: 108673. |
| [53] |
Zhang B, Zhang C, Tang RJ, et al. Two magnesium transporters in the chloroplast inner envelope essential for thylakoid biogenesis in Arabidopsis[J]. New Phytol, 2022, 236(2): 464-478.
doi: 10.1111/nph.18349 pmid: 35776059 |
| [1] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| [2] | 童玮婧, 罗数, 陆新露, 沈建福, 陆柏益, 李开绵, 马秋香, 张鹏. CRISPR/Cas9编辑MeHNL基因创制低生氰糖苷木薯[J]. 生物技术通报, 2024, 40(9): 11-19. |
| [3] | 满全财, 孟姿诺, 李伟, 蔡心汝, 苏润东, 付长青, 高顺娟, 崔江慧. 马铃薯AQP基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 51-63. |
| [4] | 赵平娟, 林晨俞, 王梦月, 张秀春, 李淑霞, 阮孟斌. 木薯SDH蛋白的序列分析及其与MeH1.2关系的研究[J]. 生物技术通报, 2024, 40(9): 74-81. |
| [5] | 吴娟, 武小娟, 王沛捷, 谢锐, 聂虎帅, 李楠, 马艳红. 彩色马铃薯花青素合成相关ERF基因筛选及表达分析[J]. 生物技术通报, 2024, 40(9): 82-91. |
| [6] | 武帅, 辛燕妮, 买春海, 穆晓娅, 王敏, 岳爱琴, 赵晋忠, 吴慎杰, 杜维俊, 王利祥. 大豆GS基因家族全基因组鉴定及胁迫响应分析[J]. 生物技术通报, 2024, 40(8): 63-73. |
| [7] | 杨巍, 赵丽芬, 唐兵, 周麟笔, 杨娟, 莫传园, 张宝会, 李飞, 阮松林, 邓英. 芥菜SRO基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 129-141. |
| [8] | 周麟, 黄顺满, 苏文坤, 姚响, 屈燕. 滇山茶bHLH基因家族鉴定及花色形成相关基因筛选[J]. 生物技术通报, 2024, 40(8): 142-151. |
| [9] | 张明亚, 庞胜群, 刘玉东, 苏永峰, 牛博文, 韩琼琼. 番茄FAD基因家族的鉴定与表达分析[J]. 生物技术通报, 2024, 40(7): 150-162. |
| [10] | 臧文蕊, 马明, 砗根, 哈斯阿古拉. 甜瓜BZR转录因子家族基因的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(7): 163-171. |
| [11] | 王健, 杨莎, 孙庆文, 陈宏宇, 杨涛, 黄园. 金钗石斛bHLH转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(6): 203-218. |
| [12] | 李梦然, 叶伟, 李赛妮, 张维阳, 李建军, 章卫民. Lithocarols类化合物生物合成基因litI的表达及其启动子功能分析[J]. 生物技术通报, 2024, 40(6): 310-318. |
| [13] | 胡永波, 雷雨田, 杨永森, 陈馨, 林黄昉, 林碧英, 刘爽, 毕格, 申宝营. 黄瓜和南瓜Bcl-2相关抗凋亡家族全基因组鉴定与表达模式分析[J]. 生物技术通报, 2024, 40(6): 219-237. |
| [14] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [15] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||