








生物技术通报 ›› 2025, Vol. 41 ›› Issue (7): 336-346.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0008
王辉1,2( ), 范灵熙2, 孙纪录1, 王苑3, 伍宁丰2, 田健3, 关菲菲2(
), 范灵熙2, 孙纪录1, 王苑3, 伍宁丰2, 田健3, 关菲菲2( )
)
收稿日期:2025-01-04
出版日期:2025-07-26
发布日期:2025-07-22
通讯作者:
关菲菲,女,博士,副研究员,研究方向 :酶蛋白分子设计及改造;E-mail: guanfeifei@caas.cn作者简介:王辉,男,硕士研究生,研究方向 :酶蛋白分子设计及改造;E-mail: asherhui0120@163.com
基金资助:
WANG Hui1,2( ), FAN Ling-xi2, SUN Ji-lu1, WANG Yuan3, WU Ning-feng2, TIAN Jian3, GUAN Fei-fei2(
), FAN Ling-xi2, SUN Ji-lu1, WANG Yuan3, WU Ning-feng2, TIAN Jian3, GUAN Fei-fei2( )
)
Received:2025-01-04
Published:2025-07-26
Online:2025-07-22
摘要:
目的 溶菌酶可作为抑菌剂而广泛应用于食品、生物、医药等领域。但溶菌酶作为一种生物活性物质,其稳定性受温度影响较大,难以满足不同行业的需求。因此采用一种结合人工智能模型设计并筛选蛋白质突变体的改造策略,提高溶菌酶的热稳定性,扩大溶菌酶的实际应用范围。 方法 研究材料为瘤胃原虫基因组来源的溶菌酶RPL187,通过大肠杆菌异源表达蛋白质进行后续实验,利用国标法检测溶菌酶RPL187在不同温度(37、45、50、55 ℃)处理不同时间(0、1、2、4、8 h)下剩余酶比活的变化情况;基于人工智能模型生成并筛选RPL187的多点突变体,同样利用国标法检测溶菌酶突变体在不同温度处理不同时间下剩余酶比活的变化情况,并通过测定野生型与突变体Tm值、自由能、氢键数量、二级结构含量等变化进而研究热稳定性提高的机制。 结果 RPL187在37 ℃和pH 6.5条件下的酶比活为(142 000±2 000)U/mg,比蛋清溶菌酶高5倍;RPL187在37 ℃和45 ℃条件下较稳定,但随着温度升高和热处理时间延长,酶比活出现明显下降的情况,在55 ℃条件下孵育1 h,酶比活下降88%左右;为了提高RPL187的热稳定性,基于人工智能模型共筛选11个RPL187多点突变体;有7个突变体成功在大肠杆菌中可溶性表达,其中RPL187-592和RPL187-209具有抑制藤黄微球菌的活性;进一步热稳定性的检测结果显示,RPL187-592和RPL187-209在50 ℃下经热处理8 h后剩余酶比活较野生型分别高4.43倍和2.29倍,Tm值较野生型分别提高2.06 ℃和2.41 ℃,并且自由能较野生型分别降低1.57 kcal/mol和0.43 kcal/mol,表现出较野生型更稳定的构象和更高的热稳定性。与野生型相比,突变体RPL187-592的分子内氢键增加了3个,且氨基酸由亲水向疏水的转变(K2V和K137V)均有助于提高蛋白质的热稳定性;而在RPL187-209中,可能是由于氨基酸由柔性向刚性(K78P和K108P)以及由亲水向疏水(K137A)的转变而增加热稳定性。 结论 本改造策略可以有效提高蛋白质热稳定性,对于扩大溶菌酶应用范围的实际需求有着重要意义,同时也为相关研究提供可参考的依据。
王辉, 范灵熙, 孙纪录, 王苑, 伍宁丰, 田健, 关菲菲. 基于蛋白智能模型提升溶菌酶RPL187的热稳定性[J]. 生物技术通报, 2025, 41(7): 336-346.
WANG Hui, FAN Ling-xi, SUN Ji-lu, WANG Yuan, WU Ning-feng, TIAN Jian, GUAN Fei-fei. Enhancing the Thermostability of Lysozyme RPL187 Based on Protein Intelligence Models[J]. Biotechnology Bulletin, 2025, 41(7): 336-346.

图1 RPL187表达与活性测定A: SDS-PAGE蛋白凝胶电泳图,粗酶液为诱导后破碎产生的杂蛋白,纯酶液为经纯化获得的纯蛋白;B: 粗酶液牛津杯抑菌圈实验图,NC为阴性对照,PC为阳性对照,CK为空白对照;C: 国标法检测溶菌RPL187酶比活图,NC为阴性对照,PC为阳性对照。****P<0.000 1
Fig. 1 Expression and activity assay of RPL187A: SDS-PAGE protein gel electrophoresis, crude enzyme solution was the heterogeneous protein produced by fragmentation after induction, and pure enzyme solution was obtained by purification. B: Oxford cup circle of inhibition experiment graph of crude enzyme solution, NC is the negative control, PC is the positive control, and CK is the blank control. C: Specific activity graph of lysin RPL187 enzyme detected by the national standard method, NC is the negative control, and PC is the positive control. ****P<0.000 1
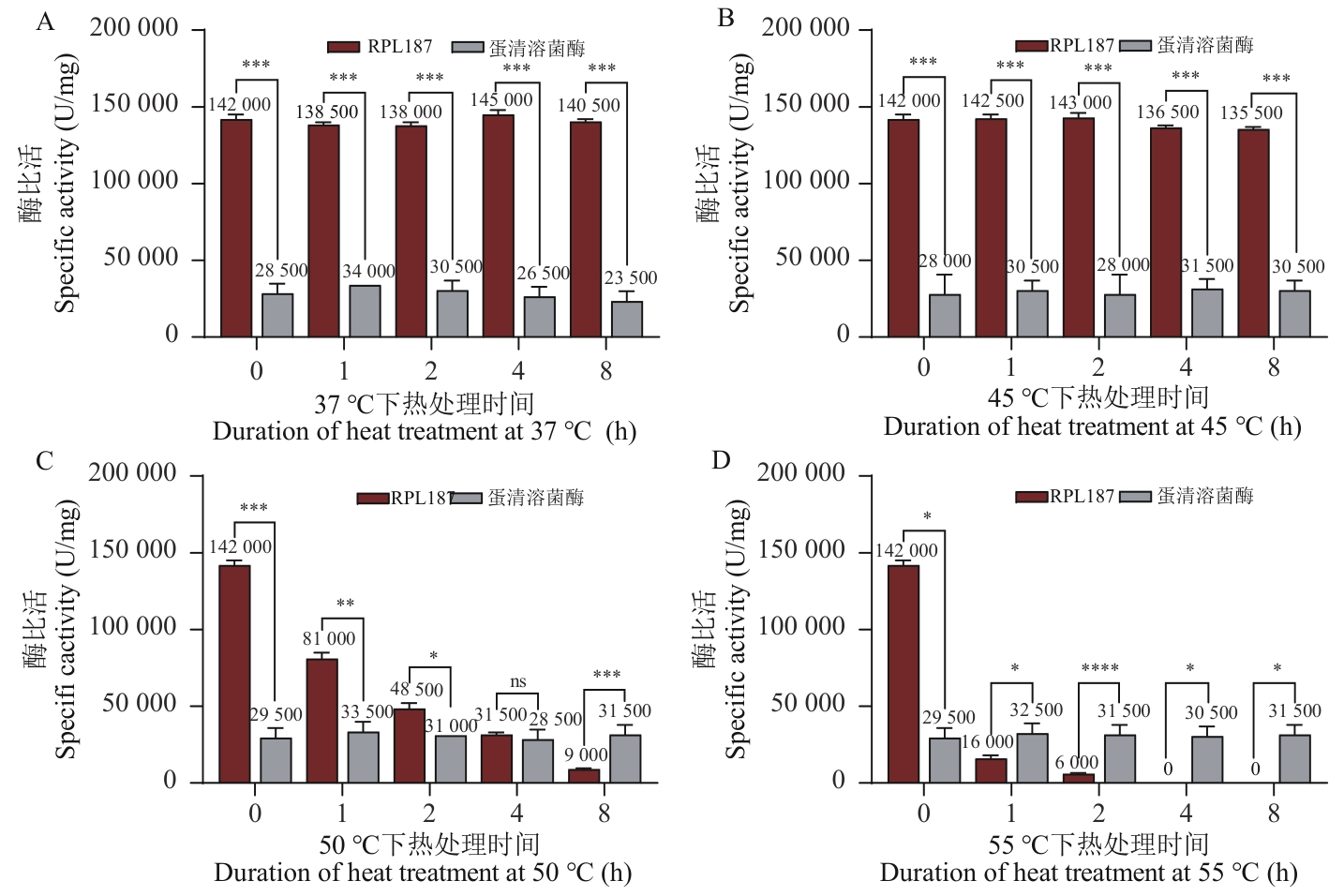
图2 国标法测定溶菌酶RPL187热稳定性A:37 ℃下不同热处理时间的溶菌酶RPL187和蛋清溶菌酶HEWL酶比活变化图;B:45 ℃下不同热处理时间的溶菌酶RPL187和蛋清溶菌酶HEWL酶比活变化图;C:50 ℃下不同热处理时间的溶菌酶RPL187和蛋清溶菌酶HEWL酶比活变化图;D:55 ℃下不同热处理时间的溶菌酶RPL187和蛋清溶菌酶HEWL酶比活变化图。ns: P≥0.05; *P<0.05; **P<0.01; ***P<0.001; ****P<0.000 1。下同
Fig. 2 Thermostability of lysozyme RPL187 by national standard methodA: Specific activity of lysozyme RPL187 and egg white lysozyme HEWL at different durations of heat treatment at 37 ℃. B: Specific activity of lysozyme RPL187 and egg white lysozyme HEWL at different durations of heat treatment at 45 ℃. C: Specific activity of lysozyme RPL187 and egg white lysozyme HEWL at different durations of heat treatment at 50 ℃. D: Specific activity of lysozyme RPL187 and egg white lysozyme at different durations of heat treatment at 55 ℃. ns: P≥0.05; *P<0.05; **P<0.01; ***P<0.001; ****P<0.000 1. The same below

图3 RPL187突变体的表达与活力检测A: RPL187及突变体粗酶液SDS-PAGE蛋白凝胶电泳图;B: 粗酶液牛津杯抑菌圈实验图
Fig. 3 Expression and viability assay of the RPL187 mutantsA: SDS-PAGE protein gel electrophoresis of crude enzyme solution of RPL187 and mutants. B: Oxford cup circle of inhibition experiment graph of crude enzyme solution

图4 国标法测定RPL187及突变体的热稳定性A:50 ℃不同热处理时间的蛋清溶菌酶、RPL187及突变体RPL187-209和RPL187-592的酶比活变化图;B:55 ℃不同热处理时间的蛋清溶菌酶、RPL187及突变体RPL187-209和RPL187-592的酶比活变化图
Fig. 4 Thermostability of RPL187 and mutants by national standard methodA: Plots of changes in specific activity of egg white lysozyme, RPL187 and mutants RPL187-209 and RPL187-592 at different durations of heat treatment at 50 ℃. B: Plots of changes in specific activity of egg white lysozyme, RPL187 and mutants RPL187-209 and RPL187-592 at different durations of heat treatment at 55 ℃
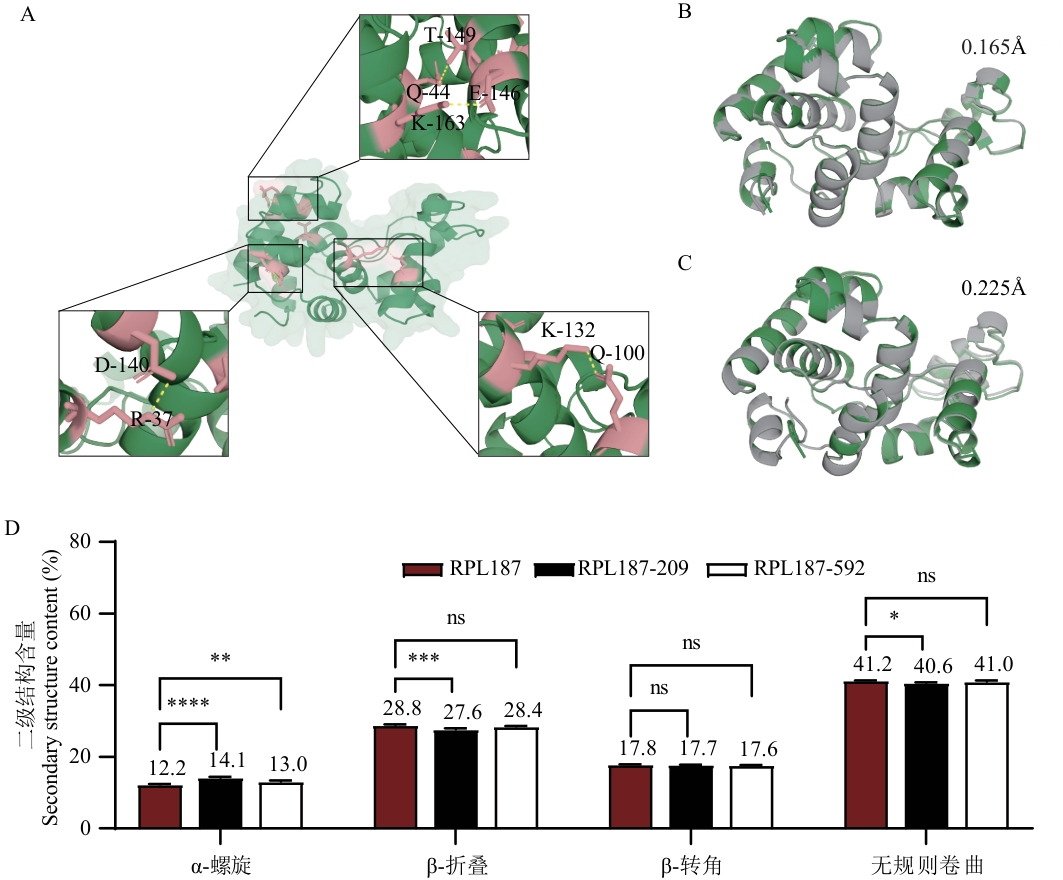
图5 热稳定性提高的机制分析图A:Pymol可视化展示突变体RPL187-592相对于野生型增加的氢键图;B:利用Pymol计算突变体RPL187-209与野生型RPL187的RMSD (root mean square deviation) 值;C:利用Pymol计算突变体RPL187-592与野生型RPL187的RMSD值;基于Pymol绘制的突变体(灰色)与野生型(绿色)的蛋白结构叠合图,直观展示突变引起的构象变化;RMSD(均方根偏差)用于衡量两个蛋白质结构之间的相似性,值越小表示结构差异越小;D:利用圆二色谱仪(CD)检测野生型RPL187及突变体RPL187-209、RPL187-592的二级结构含量变化图
Fig. 5 Analysis diagram of the mechanism of improving thermostabilityA: Pymol visualisation showing the increased hydrogen bonding map of mutant RPL187-592 related to the wild type. B: Calculating RMSD (root mean square deviation) values of mutant RPL187-209 versus wild type RPL187 using Pymol. C: Calculating RMSD values of mutant RPL187-592 versus wild type RPL187 using Pymol. Based on the Pymol (grey) and wild-type (green) protein structure superimposed diagrams to visually demonstrate the conformational changes caused by the mutation; RMSD was used to measure the similarity between the structures of the two proteins, with smaller values indicating smaller structural differences. D: Detection of the secondary structure content changes of the wild-type RPL187 and the mutants RPL187-209 and RPL187-592 by circular dichroism (CD)
名称 Name | 模型性能 评估值 Evaluated value of model performance | Tm值 预测模型 Tm value predicting model | 最适生长温度 预测模型 Predicting model for optimal growth temperature | 大肠杆菌 表达量预测模型 Predicting model for E. coli expression | 枯草芽孢杆菌 表达量预测模型 Predicting model for B. subtilis expression | 酿酒酵母 表达量预测模型 Predicting model for S. cerevisiae expression | 总和 Sum |
|---|---|---|---|---|---|---|---|
| RPL187-592 | 0.83 | -0.69 | 2.42 | 2.08 | 1.32 | 1.60 | 6.72 |
| RPL187-319 | 0.80 | 1.26 | 1.28 | 1.18 | 0.60 | 1.79 | 6.10 |
| RPL187-496 | 0.80 | 0.49 | 0.89 | 1.98 | 0.34 | 2.18 | 5.88 |
| RPL187-622 | 0.80 | 0.13 | -0.25 | 2.31 | 0.81 | 2.63 | 5.63 |
| RPL187-168 | 0.84 | 2.15 | 1.96 | -0.65 | 1.70 | -1.34 | 3.82 |
| RPL187-857 | 0.82 | 0.95 | 1.78 | -0.87 | 1.92 | -0.38 | 3.40 |
| RPL187-649 | 0.84 | 1.69 | 1.20 | -0.65 | 1.29 | -1.02 | 2.50 |
| RPL187-954 | 0.83 | 1.09 | 0.43 | -0.54 | 0.11 | 0.27 | 1.37 |
| RPL187-209 | 0.80 | -0.96 | -0.34 | 1.71 | 0.64 | -0.01 | 1.05 |
| RPL187-963 | 0.83 | 0.22 | 0.33 | 0.18 | 0.04 | -0.27 | 0.51 |
| RPL187-700 | 0.82 | -0.93 | 0.86 | -0.60 | 0.82 | -0.12 | 0.03 |
| RPL187 | -1.00 | -1.37 | -1.25 | -1.12 | -1.58 | -1.35 | -6.67 |
表1 多模型结合筛选RPL187突变体评价分值表
Table 1 Multi-model combined screening of RPL187 mutant evaluation score sheet
名称 Name | 模型性能 评估值 Evaluated value of model performance | Tm值 预测模型 Tm value predicting model | 最适生长温度 预测模型 Predicting model for optimal growth temperature | 大肠杆菌 表达量预测模型 Predicting model for E. coli expression | 枯草芽孢杆菌 表达量预测模型 Predicting model for B. subtilis expression | 酿酒酵母 表达量预测模型 Predicting model for S. cerevisiae expression | 总和 Sum |
|---|---|---|---|---|---|---|---|
| RPL187-592 | 0.83 | -0.69 | 2.42 | 2.08 | 1.32 | 1.60 | 6.72 |
| RPL187-319 | 0.80 | 1.26 | 1.28 | 1.18 | 0.60 | 1.79 | 6.10 |
| RPL187-496 | 0.80 | 0.49 | 0.89 | 1.98 | 0.34 | 2.18 | 5.88 |
| RPL187-622 | 0.80 | 0.13 | -0.25 | 2.31 | 0.81 | 2.63 | 5.63 |
| RPL187-168 | 0.84 | 2.15 | 1.96 | -0.65 | 1.70 | -1.34 | 3.82 |
| RPL187-857 | 0.82 | 0.95 | 1.78 | -0.87 | 1.92 | -0.38 | 3.40 |
| RPL187-649 | 0.84 | 1.69 | 1.20 | -0.65 | 1.29 | -1.02 | 2.50 |
| RPL187-954 | 0.83 | 1.09 | 0.43 | -0.54 | 0.11 | 0.27 | 1.37 |
| RPL187-209 | 0.80 | -0.96 | -0.34 | 1.71 | 0.64 | -0.01 | 1.05 |
| RPL187-963 | 0.83 | 0.22 | 0.33 | 0.18 | 0.04 | -0.27 | 0.51 |
| RPL187-700 | 0.82 | -0.93 | 0.86 | -0.60 | 0.82 | -0.12 | 0.03 |
| RPL187 | -1.00 | -1.37 | -1.25 | -1.12 | -1.58 | -1.35 | -6.67 |
名称 Name | 突变数 Number of mutations | 突变氨基酸 Mutated amino acids |
|---|---|---|
| RPL187-592 | 6 | K2V__C26A__A43S__S115D__K137A__K150R |
| RPL187-319 | 8 | N3R__Q11A__C26A__Y54T__S65A__I148V__V152I__F173I |
| RPL187-496 | 4 | C26A__Y54T__K78P__C82R |
| RPL187-622 | 5 | M10L__C26A__C82R__Y89L__M111Q |
| RPL187-168 | 6 | V5I__R39A__L51S__F84Y__R109N__E146T |
| RPL187-857 | 7 | V5I__K12A__R53Q__Q56R__S65A__K137A__E146T |
| RPL187-649 | 5 | V5I__M10L__M13L__S118A__K132S |
| RPL187-954 | 7 | Q11A__R39A__A50G__L51S__S65A__F84Y__K137A |
| RPL187-209 | 4 | K78P__Q99T__K108P__K137A |
| RPL187-963 | 3 | M10L__R39A__K132S |
| RPL187-700 | 8 | K78P__A87R__Y89L__M111Q__S118A__K132S__E146T__F173I |
| RPL187 | 0 | - |
表2 RPL187稳定性改造突变体的突变位点变化表
Table 2 Table of mutation site changes in RPL187 stability-modified mutants
名称 Name | 突变数 Number of mutations | 突变氨基酸 Mutated amino acids |
|---|---|---|
| RPL187-592 | 6 | K2V__C26A__A43S__S115D__K137A__K150R |
| RPL187-319 | 8 | N3R__Q11A__C26A__Y54T__S65A__I148V__V152I__F173I |
| RPL187-496 | 4 | C26A__Y54T__K78P__C82R |
| RPL187-622 | 5 | M10L__C26A__C82R__Y89L__M111Q |
| RPL187-168 | 6 | V5I__R39A__L51S__F84Y__R109N__E146T |
| RPL187-857 | 7 | V5I__K12A__R53Q__Q56R__S65A__K137A__E146T |
| RPL187-649 | 5 | V5I__M10L__M13L__S118A__K132S |
| RPL187-954 | 7 | Q11A__R39A__A50G__L51S__S65A__F84Y__K137A |
| RPL187-209 | 4 | K78P__Q99T__K108P__K137A |
| RPL187-963 | 3 | M10L__R39A__K132S |
| RPL187-700 | 8 | K78P__A87R__Y89L__M111Q__S118A__K132S__E146T__F173I |
| RPL187 | 0 | - |
名称 Name | 蛋白质熔化温度 Tm(℃) | 自由能变化 Free energy change(kcal/mol) |
|---|---|---|
| RPL187 | 52.62±0.26 | 0 |
| RPL187-209 | 54.68±0.28 | -1.57 |
| RPL187-592 | 55.03±0.28 | -0.43 |
表3 RPL187与突变体的Tm值与自由能变化
Table 3 Changes in Tm values and free energy changes in RPL187 and mutants
名称 Name | 蛋白质熔化温度 Tm(℃) | 自由能变化 Free energy change(kcal/mol) |
|---|---|---|
| RPL187 | 52.62±0.26 | 0 |
| RPL187-209 | 54.68±0.28 | -1.57 |
| RPL187-592 | 55.03±0.28 | -0.43 |
| [1] | Wu TT, Jiang QQ, Wu D, et al. What is new in lysozyme research and its application in food industry? A review [J]. Food Chem, 2019, 274: 698-709. |
| [2] | Abdou AM, Higashiguchi S, Aboueleinin AM, et al. Antimicrobial peptides derived from hen egg lysozyme with inhibitory effect against Bacillus species [J]. Food Contr, 2007, 18(2): 173-178. |
| [3] | Abey SK, Yuana Y, Joseph PV, et al. Lysozyme association with circulating RNA, extracellular vesicles, and chronic stress [J]. BBA Clin, 2017, 7: 23-35. |
| [4] | Bahrami A, Delshadi R, Assadpour E, et al. Antimicrobial-loaded nanocarriers for food packaging applications [J]. Adv Colloid Interface Sci, 2020, 278: 102140. |
| [5] | Wu HY, Cao DN, Liu TX, et al. Purification and characterization of recombinant human lysozyme from eggs of transgenic chickens [J]. PLoS One, 2015, 10(12): e0146032. |
| [6] | Jiang MF, Hu MJ, Ren HH, et al. Molecular cloning and characterization of a new C-type lysozyme gene from yak mammary tissue [J]. Asian-Australas J Anim Sci, 2015, 28(12): 1774-1783. |
| [7] | Kim YK, Nam YK. Molecular characterization and expression pattern of c-type and g-type lysozyme isoforms in starry flounder Platichthys stellate infected with Streptococcus parauberis [J]. Fish Sci, 2015, 81: 353-363. |
| [8] | Shahmohammadi A. Lysozyme separation from chicken egg white: a review [J]. Eur Food Res Technol, 2018, 244(4): 577-593. |
| [9] | Gill AO, Holley RA. Inhibition of bacterial growth on ham and bologna by lysozyme, nisin and EDTA [J]. Food Res Int, 2000, 33(2): 83-90. |
| [10] | Ferraboschi P, Ciceri S, Grisenti P. Applications of lysozyme, an innate immune defense factor, as an alternative antibiotic [J]. Antibiotics, 2021, 10(12): 1534. |
| [11] | Ercan D, Demirci A. Recent advances for the production and recovery methods of lysozyme [J]. Crit Rev Biotechnol, 2016, 36(6): 1078-1088. |
| [12] | Salazar O, Asenjo JA. Enzymatic Lysis of microbial cells [J]. Biotechnol Lett, 2007, 29(7): 985-994. |
| [13] | Wells JE, Berry ED, Kalchayanand N, et al. Effect of lysozyme or antibiotics on faecal zoonotic pathogens in nursery pigs [J]. J Appl Microbiol, 2015, 118(6): 1489-1497. |
| [14] | Nawaz N, Wen S, Wang FH, et al. Lysozyme and its application as antibacterial agent in food industry [J]. Molecules, 2022, 27(19): 6305. |
| [15] | Elkordy AA, Forbes RT, Barry BW. Stability of crystallised and spray-dried lysozyme [J]. Int J Pharm, 2004, 278(2): 209-219. |
| [16] | 唐汉颖, 韦加娜. 新型抗菌纸的制备及其抗菌性能研究 [C]. 中国化学会, 2016:198-199. |
| Tang HY, Wei JN. Preparation of new antimicrobial paper and research on its antimicrobial properties [C]. Chinese Chemical Society, 2016: 198-199. | |
| [17] | 田健, 王平, 伍宁丰, 等. 理性设计提高蛋白质热稳定性的研究进展 [J]. 生物技术进展, 2012, 2(4): 233-239. |
| Tian J, Wang P, Wu NF, et al. Recent advances in the rational design to improve the protein thermostability [J]. Curr Biotechnol, 2012, 2(4): 233-239. | |
| [18] | Abdulkareem RA, Doekhie A, Fotaki N, et al. Thermal stabilisation of lysozyme through ensilication [J]. Molecules, 2024, 29(17): 4207. |
| [19] | Venkataramani S, Truntzer J, Coleman DR. Thermal stability of high concentration lysozyme across varying pH: a Fourier Transform Infrared study [J]. J Pharm Bioallied Sci, 2013, 5(2): 148-153. |
| [20] | Avanti C, Saluja V, van Streun ELP, et al. Stability of lysozyme in aqueous extremolyte solutions during heat shock and accelerated thermal conditions [J]. PLoS One, 2014, 9(1): e86244. |
| [21] | Steven Johnson L, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure [J]. BMC Bioinformatics, 2010, 11: 431. |
| [22] | Liu TY, Gao H, Ren XP, et al. Protein-protein interaction and site prediction using transfer learning [J]. Brief Bioinform, 2023, 24(6): bbad376. |
| [23] | Schymkowitz J, Borg J, Stricher F, et al. The FoldX web server: an online force field [J]. Nucleic Acids Res, 2005, 33(Web Server issue): W382-W388. |
| [24] | Kalayan J, Chakravorty A, Warwicker J, et al. Total free energy analysis of fully hydrated proteins [J]. Proteins, 2023, 91(1): 74-90. |
| [25] | Sun ZT, Liu Q, Qu G, et al. Utility of B-factors in protein science: interpreting rigidity, flexibility, and internal motion and engineering thermostability [J]. Chem Rev, 2019, 119(3): 1626-1665. |
| [26] | Britton KL, Baker PJ, Borges KM, et al. Insights into thermal stability from a comparison of the glutamate dehydrogenases from Pyrococcus furiosus and Thermococcus litoralis [J]. Eur J Biochem, 1995, 229(3): 688-695. |
| [27] | Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold [J]. Nature, 2021, 596(7873): 583-589. |
| [28] | Ferruz N, Schmidt S, Höcker B. ProteinTools: a toolkit to analyze protein structures [J]. Nucleic Acids Res, 2021, 49(W1): W559-W566. |
| [29] | Pace CN, Fu HL, Lee Fryar K, et al. Contribution of hydrogen bonds to protein stability [J]. Protein Sci, 2014, 23(5): 652-661. |
| [30] | Ma BH, Liu DH, Zheng MJ, et al. Development of a double-stapled peptide stabilizing both α-helix and β-sheet structures for degrading transcription factor AR-V7 [J]. JACS Au, 2024, 4(2): 816-827. |
| [31] | Burgos MI, Ochoa A, Perillo MA. β-sheet to α-helix conversion and thermal stability of β-Galactosidase encapsulated in a nanoporous silica gel [J]. Biochem Biophys Res Commun, 2019, 508(1): 270-274. |
| [32] | Yu M, Silva TC, van Opstal A, et al. The investigation of protein diffusion via H-cell microfluidics [J]. Biophys J, 2019, 116(4): 595-609. |
| [33] | Clegg JR, Peppas NA. Design of synthetic hydrogel compositions for noncovalent protein recognition [J]. ACS Appl Mater Interfaces, 2023. |
| [34] | Ho TC, Chang CC, Chan HP, et al. Hydrogels: properties and applications in biomedicine [J]. Molecules, 2022, 27(9): 2902. |
| [35] | Levashov PA, Matolygina DA, Ovchinnikova ED, et al. The bacteriolytic activity of native and covalently immobilized lysozyme against Gram-positive and Gram-negative bacteria is differentially affected by charged amino acids and glycine [J]. FEBS Open Bio, 2019, 9(3): 510-518. |
| [36] | Carmen Salinas-Garcia M, Plaza-Garrido M, Alba-Elena D, et al. Major conformational changes in the structure of lysozyme obtained from a crystal with a very low solvent content [J]. Acta Crystallogr F Struct Biol Commun, 2019, 75(Pt 11): 687-696. |
| [37] | Pal S, Mitra RK. Nonpolar hydrophobic amino acids tune the enzymatic activity of lysozyme [J]. Biophys Chem, 2022, 288: 106842. |
| [38] | Funahashi J, Takano K, Yamagata Y, et al. Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions [J]. Biochemistry, 2000, 39(47): 14448-14456. |
| [1] | 车建美, 郑雪芳, 王阶平, 陈燕萍, 陈冰星, 刘波. 一株产纤维素酶菌株的筛选、鉴定及全基因组分析[J]. 生物技术通报, 2025, 41(3): 294-307. |
| [2] | 杨悦, 陶妍, 谢晶, 钱韻芳. 基于重组毕赤酵母的草鱼C型溶菌酶生物合成及其抑菌活性[J]. 生物技术通报, 2021, 37(12): 169-179. |
| [3] | 张春晨, 胡双艳, 阮海华. 人源溶菌酶在大肠杆菌中的表达与复性研究[J]. 生物技术通报, 2020, 36(3): 153-161. |
| [4] | 王强厚, 陶妍, 崔旭, 颜倩倩, 张亚莉. 三疣梭子蟹C型溶菌酶在毕赤酵母中的表达及其抑菌活性[J]. 生物技术通报, 2018, 34(10): 135-142. |
| [5] | 冯亚东,陶妍,李雯,崔旭,王强厚. 斑点叉尾鮰C型溶菌酶在毕赤酵母中的表达及其抑菌活性[J]. 生物技术通报, 2017, 33(7): 195-202. |
| [6] | 闻崇炜,赵烨清,石莉,欧阳臻. 聚乙二醇沉淀联用双水相萃取法纯化蛋清溶菌酶的研究[J]. 生物技术通报, 2017, 33(5): 89-93. |
| [7] | 王赟, 翟素珍, 张春林, 王吉平. 美洲大蠊i型溶菌酶的原核表达及多克隆抗体制备[J]. 生物技术通报, 2016, 32(1): 138-143. |
| [8] | 彭传林, 魏川川, 吴建伟, 王宇, 修江帆, 尚小丽, 赵学军. 家蝇抗真菌肽-溶菌酶基因的克隆、表达及序列分析[J]. 生物技术通报, 2015, 31(5): 200-205. |
| [9] | 孙璐, 刘志文, 邹丹, 李丹, 潘博, 丛丽娜. 海参溶菌酶枯草芽孢杆菌基因工程菌构建[J]. 生物技术通报, 2014, 30(6): 150-154. |
| [10] | 李丹, 李成, 孙璐, 邹丹, 刘志文, 丛丽娜. 海参溶菌酶基因在枯草芽孢杆菌WB600中的整合及表达[J]. 生物技术通报, 2014, 30(4): 139-146. |
| [11] | 汪涛, 余辉, 梅钧, 李兴暖, 周小鸥, 赵志军. 猪链球菌2型MRP和EF双基因共表达重组腺病毒载体的构建[J]. 生物技术通报, 2013, 0(8): 130-134. |
| [12] | 瞿兰;叶星;田园园;张莉莉;高风英;卢迈新;张蕊;. 罗非鱼3种C型溶菌酶重组蛋白的制备及与几种鱼虾溶菌酶溶菌谱的比较[J]. , 2012, 0(11): 161-166. |
| [13] | 杨杰;丛丽娜;夏涛;常艺海;栾桂花;. 日本对虾c型溶菌酶的高效重组表达及产物分析[J]. , 2012, 0(06): 99-105. |
| [14] | 张清华;夏雪琴;克力比努尔·热合曼;李江伟;. 新疆双峰驼天然单域抗体库的构建及初步鉴定[J]. , 2011, 0(10): 151-155. |
| [15] | 王艳玲;李东;王秀利;. 海参免疫相关基因的研究进展[J]. , 2011, 0(09): 22-26. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||