








生物技术通报 ›› 2025, Vol. 41 ›› Issue (9): 147-158.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0210
• 研究报告 • 上一篇
收稿日期:2025-02-28
出版日期:2025-09-26
发布日期:2025-09-24
通讯作者:
尹晓,男,博士,副教授,研究方向 :果树育种与生物技术;E-mail: yinxiao90@nxu.edu.cn作者简介:李珊,女,硕士研究生,研究方向 :果树育种与生物技术;E-mail: lshanym@163.com
基金资助:
LI Shan( ), MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao(
), MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao( )
)
Received:2025-02-28
Published:2025-09-26
Online:2025-09-24
摘要:
目的 研究葡萄(Vitis vinifera)S期激酶相关蛋白1(S-phase kinase-associated-protein 1, SKP1)基因家族(VvSKP1)特征和表达分析,为后续深入研究VvSKP1基因家族的生物学功能提供理论依据。 方法 基于葡萄基因组数据,利用生物信息学方法对SKP1基因家族成员进行筛选鉴定,对其编码蛋白质的特征、系统进化关系、基因结构、染色体定位、顺式作用元件以及共线性关系等进行分析,并利用实时荧光定量PCR(RT-qPCR)分析其在灰霉菌和霜霉菌侵染下的表达模式。 结果 从葡萄基因组内共鉴定到11个SKP1家族基因(VvSKP1-1‒VvSKP1-11),分布于7条染色体和1条未知染色体上,编码119个(VvSKP1-3)‒438(VvSKP1-8)个氨基酸。系统发育分析显示,SKP1家族蛋白可分为3个亚家族,葡萄的11个SKP1蛋白只分布在第I和第Ⅱ亚家族,第III亚家族全是小麦SKP1基因家族成员。同亚家族的VvSKP1成员在基因结构和保守基序上表现出一定的相似性。基因的启动子顺式作用元件主要与光响应、激素响应和胁迫应答有关。共线性分析发现VvSKP1有8对同源基因。选择压力分析表明,VvSKP1基因处于纯化选择。RT-qPCR分析表明,分别有11和10个VvSKP1基因积极响应灰霉菌和霜霉菌的侵染。 结论 从葡萄基因组中鉴定出11个VvSKP1s基因家族成员,该家族成员在灰霉病和霜霉病胁迫下的表达模式存在差异,表明葡萄SKP1基因家族可能参与葡萄灰霉病和霜霉病胁迫响应过程。
李珊, 马登辉, 马红义, 姚文孔, 尹晓. 葡萄SKP1基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(9): 147-158.
LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.)[J]. Biotechnology Bulletin, 2025, 41(9): 147-158.
| 基因 Gene | 上游引物 Forward primer (5′‒3′) | 下游引物 Reverse primer (5′‒3′) |
|---|---|---|
| VvSKP1-1 | TGCTCTCCAGTCCAGAACCATCC | AATCGGTGTTGAGATCAATCAAGTGTTTC |
| VvSKP1-2 | TGATGATCCACGGATTCGGCTATTG | ACGCTCCTCTTCAACCTCAACATTC |
| VvSKP1-3 | ACAATCCTGGATATTCTACGAGCTTCTG | ACCAAATAAATCTCTCACCTCTTCTACCG |
| VvSKP1-4 | GAAGGAGGTGGATGAGATGAAGAAGTG | AATCAACCCTGCTATGCTCAAATAATTGG |
| VvSKP1-5 | AGCAAACTATCTGAACATCAAGAGTCTCC | CCGCCGCACCTCCTCCTC |
| VvSKP1-6 | GGTGAATGGTAGAACCCTGGCTAAAG | CACTTCTTCATCTCATCCACCTCCTTC |
| VvSKP1-7 | CAGCGGTATCCTTGCGATGGTG | CATGATGAGGTGGAACAGAGTAGCC |
| VvSKP1-8 | GGGCTGGGCTGCTGATTTCTTC | TTGATATGTAGATAGTCTGCTGCCTTGATG |
| VvSKP1-9 | ATCAAGTAATAGGACGCTCGAACAAGG | TGTCAGCAGCAGATGTCAACTCAC |
| VvSKP1-10 | CCGATTCTTGGCGATGGTGATAGAG | ATGATGAGATGGTAGAGAAGTGATTGGTC |
| VvSKP1-11 | GGTTGATTGATTTGACATGCGGGAAG | CCTTATCTCAGCCTCCTCCTCTACG |
| Actin | AACCCCAAGGCCAACAGAGAAAA | CACCATCACCAGAATCCAGCACA |
表1 基因荧光定量引物信息
Table 1 Information of the primers used for RT-qPCR
| 基因 Gene | 上游引物 Forward primer (5′‒3′) | 下游引物 Reverse primer (5′‒3′) |
|---|---|---|
| VvSKP1-1 | TGCTCTCCAGTCCAGAACCATCC | AATCGGTGTTGAGATCAATCAAGTGTTTC |
| VvSKP1-2 | TGATGATCCACGGATTCGGCTATTG | ACGCTCCTCTTCAACCTCAACATTC |
| VvSKP1-3 | ACAATCCTGGATATTCTACGAGCTTCTG | ACCAAATAAATCTCTCACCTCTTCTACCG |
| VvSKP1-4 | GAAGGAGGTGGATGAGATGAAGAAGTG | AATCAACCCTGCTATGCTCAAATAATTGG |
| VvSKP1-5 | AGCAAACTATCTGAACATCAAGAGTCTCC | CCGCCGCACCTCCTCCTC |
| VvSKP1-6 | GGTGAATGGTAGAACCCTGGCTAAAG | CACTTCTTCATCTCATCCACCTCCTTC |
| VvSKP1-7 | CAGCGGTATCCTTGCGATGGTG | CATGATGAGGTGGAACAGAGTAGCC |
| VvSKP1-8 | GGGCTGGGCTGCTGATTTCTTC | TTGATATGTAGATAGTCTGCTGCCTTGATG |
| VvSKP1-9 | ATCAAGTAATAGGACGCTCGAACAAGG | TGTCAGCAGCAGATGTCAACTCAC |
| VvSKP1-10 | CCGATTCTTGGCGATGGTGATAGAG | ATGATGAGATGGTAGAGAAGTGATTGGTC |
| VvSKP1-11 | GGTTGATTGATTTGACATGCGGGAAG | CCTTATCTCAGCCTCCTCCTCTACG |
| Actin | AACCCCAAGGCCAACAGAGAAAA | CACCATCACCAGAATCCAGCACA |
蛋白 Protein | 氨基酸数目 Amino acid number | CDS长度Length of CDS (bp) | 分子质量 Molecular weight (Da) | 等电点 Isoelectric point | 不稳定指数Instability index | 脂肪系数Aliphatic index | 总平均亲水性GRAVY | 亚细胞定位预测Subcellular localization | 跨膜结构 | 信号肽 Signal peptide | 结构域 Domain |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VvSKP1-1 | 151 | 456 | 17 523.08 | 4.66 | 36.05 | 95.56 | -0.227 | 细胞核 Nucleus | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-2 | 372 | 1 119 | 42 659.31 | 5.77 | 55.66 | 76.8 | -0.833 | 细胞核 Nucleus | 无 | 无 | Skp1 |
| VvSKP1-3 | 119 | 360 | 13 436.95 | 4.15 | 36.44 | 90.08 | -0.569 | 叶绿体 Chloroplast | 无 | 无 | Skp1 |
| VvSKP1-4 | 152 | 459 | 17 258.86 | 4.77 | 29.56 | 87.24 | -0.325 | 细胞质 Cytoplasm | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-5 | 156 | 471 | 17 694.94 | 4.60 | 48.33 | 83.78 | -0.529 | 细胞核 Nucleus | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-6 | 152 | 459 | 17 286.83 | 4.77 | 31.67 | 85.99 | -0.349 | 细胞质 Cytoplasm | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-7 | 141 | 426 | 15 668.21 | 5.26 | 39.89 | 107.87 | 0.149 | 细胞质 Cytoplasm | 无 | 无 | Skp1_POZ |
| VvSKP1-8 | 438 | 1 317 | 49 885.82 | 5.67 | 38.62 | 99.77 | -0.209 | 细胞质 Cytoplasm | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-9 | 351 | 1 056 | 40 552.66 | 5.25 | 51.36 | 77.75 | -0.874 | 细胞核 Nucleus | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-10 | 162 | 489 | 18 903.61 | 4.77 | 58.12 | 93.33 | -0.359 | 细胞质 Cytoplasm | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-11 | 157 | 474 | 17 522.99 | 4.75 | 29.13 | 91.91 | -0.259 | 细胞质 Cytoplasm | 无 | 无 | Skp1、Skp1_POZ |
表2 葡萄SKP1基因家族蛋白理化性质
Table 2 Physicochemical properties of grapevine SKP1 family proteins
蛋白 Protein | 氨基酸数目 Amino acid number | CDS长度Length of CDS (bp) | 分子质量 Molecular weight (Da) | 等电点 Isoelectric point | 不稳定指数Instability index | 脂肪系数Aliphatic index | 总平均亲水性GRAVY | 亚细胞定位预测Subcellular localization | 跨膜结构 | 信号肽 Signal peptide | 结构域 Domain |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VvSKP1-1 | 151 | 456 | 17 523.08 | 4.66 | 36.05 | 95.56 | -0.227 | 细胞核 Nucleus | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-2 | 372 | 1 119 | 42 659.31 | 5.77 | 55.66 | 76.8 | -0.833 | 细胞核 Nucleus | 无 | 无 | Skp1 |
| VvSKP1-3 | 119 | 360 | 13 436.95 | 4.15 | 36.44 | 90.08 | -0.569 | 叶绿体 Chloroplast | 无 | 无 | Skp1 |
| VvSKP1-4 | 152 | 459 | 17 258.86 | 4.77 | 29.56 | 87.24 | -0.325 | 细胞质 Cytoplasm | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-5 | 156 | 471 | 17 694.94 | 4.60 | 48.33 | 83.78 | -0.529 | 细胞核 Nucleus | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-6 | 152 | 459 | 17 286.83 | 4.77 | 31.67 | 85.99 | -0.349 | 细胞质 Cytoplasm | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-7 | 141 | 426 | 15 668.21 | 5.26 | 39.89 | 107.87 | 0.149 | 细胞质 Cytoplasm | 无 | 无 | Skp1_POZ |
| VvSKP1-8 | 438 | 1 317 | 49 885.82 | 5.67 | 38.62 | 99.77 | -0.209 | 细胞质 Cytoplasm | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-9 | 351 | 1 056 | 40 552.66 | 5.25 | 51.36 | 77.75 | -0.874 | 细胞核 Nucleus | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-10 | 162 | 489 | 18 903.61 | 4.77 | 58.12 | 93.33 | -0.359 | 细胞质 Cytoplasm | 无 | 无 | Skp1、Skp1_POZ |
| VvSKP1-11 | 157 | 474 | 17 522.99 | 4.75 | 29.13 | 91.91 | -0.259 | 细胞质 Cytoplasm | 无 | 无 | Skp1、Skp1_POZ |
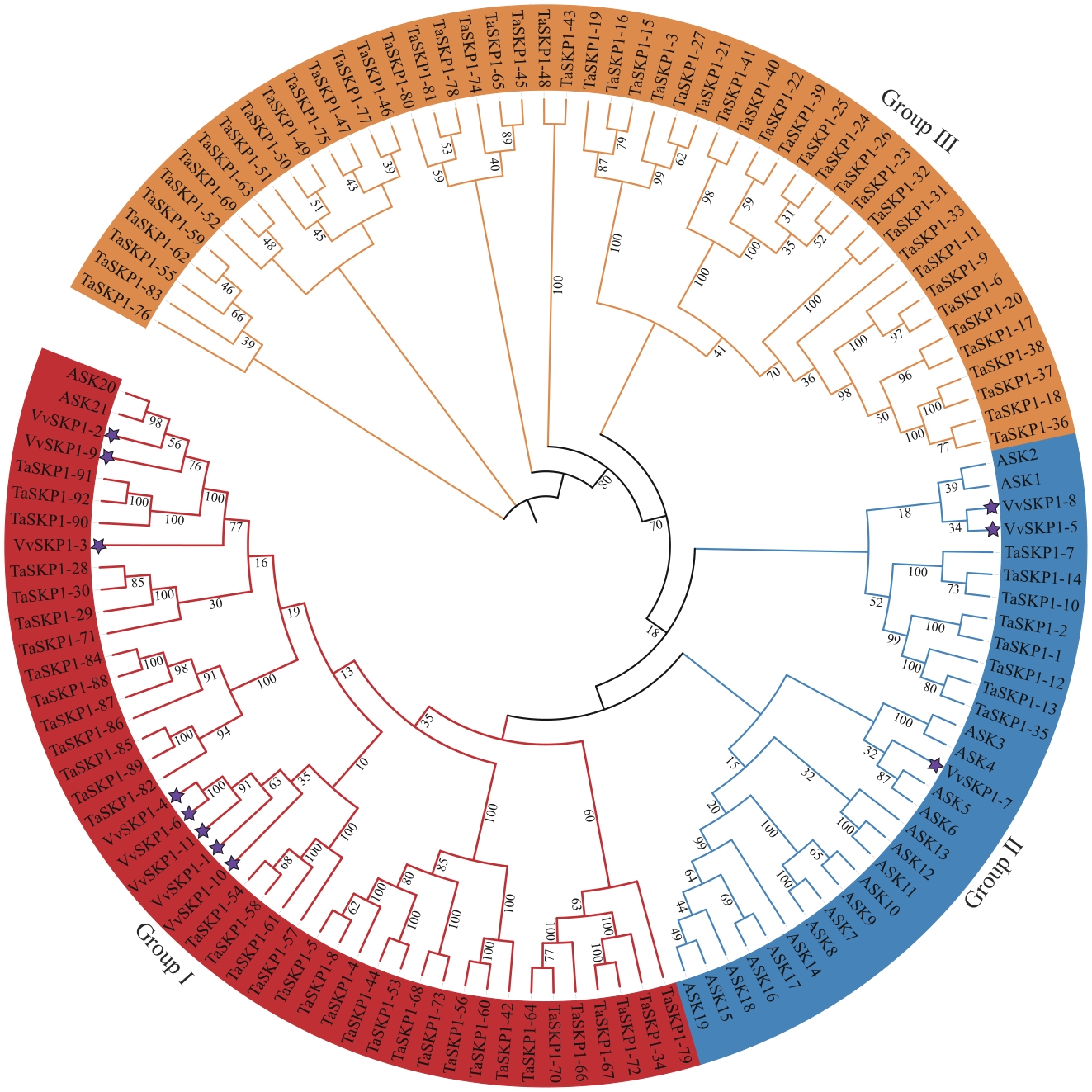
图1 葡萄(VvSKP1)、拟南芥(ASK)和小麦(TaSKP1)SKP1家族成员系统进化树紫色五角星表示葡萄SKP1家族成员
Fig. 1 Phylogenetic tree of SKP1 family members in V. vinifera (VvSKP1), A. thaliana (ASK), and T. aestivum (TaSKP1)The purple pentagram indicates the members of the SKP1 family in V. vinifera
共线性序列1 Collinearity sequence 1 | 共线性序列2 Collinearity sequence 2 | Ka | Ks | Ka/Ks | 共线性序列1 Collinearity sequence 1 | 共线性序列2 Collinearity sequence 2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|---|---|---|---|---|
| VvSKP1-1 | VvSKP1-10 | 0.484 186 | 2.676 18 | 0.180 925 | VvSKP1-11 | VvSKP1-4 | 0.318 353 | 0.978 672 | 0.325 291 |
| VvSKP1-1 | VvSKP1-11 | 0.439 432 | 2.748 94 | 0.159 855 | VvSKP1-11 | VvSKP1-5 | 0.394 356 | 3.214 360 | 0.122 685 |
| VvSKP1-1 | VvSKP1-3 | 0.744 929 | 1.804 00 | 0.412 931 | VvSKP1-11 | VvSKP1-6 | 0.312 299 | 0.961 286 | 0.324 876 |
| VvSKP1-1 | VvSKP1-4 | 0.374 420 | 3.230 87 | 0.115 888 | VvSKP1-11 | VvSKP1-7 | 0.559 368 | 2.658 880 | 0.210 377 |
| VvSKP1-1 | VvSKP1-5 | 0.401 063 | 3.083 55 | 0.130 065 | VvSKP1-3 | VvSKP1-4 | 0.657 090 | 2.075 610 | 0.316 577 |
| VvSKP1-1 | VvSKP1-6 | 0.378 107 | 3.186 02 | 0.118 677 | VvSKP1-3 | VvSKP1-5 | 0.526 860 | 2.452 040 | 0.214 866 |
| VvSKP1-1 | VvSKP1-7 | 0.551 033 | 2.400 07 | 0.229 591 | VvSKP1-3 | VvSKP1-6 | 0.674 153 | 2.036 490 | 0.331 038 |
| VvSKP1-10 | VvSKP1-11 | 0.473 489 | 2.443 63 | 0.193 765 | VvSKP1-3 | VvSKP1-7 | 0.810 727 | 1.581 060 | 0.512 774 |
| VvSKP1-10 | VvSKP1-3 | 0.639 464 | 2.206 82 | 0.289 767 | VvSKP1-4 | VvSKP1-5 | 0.364 465 | 3.387 940 | 0.107 577 |
| VvSKP1-10 | VvSKP1-4 | 0.402 901 | 2.944 32 | 0.136 840 | VvSKP1-4 | VvSKP1-6 | 0.010 264 2 | 0.014 291 7 | 0.718 192 |
| VvSKP1-10 | VvSKP1-5 | 0.384 626 | 3.164 50 | 0.121 544 | VvSKP1-4 | VvSKP1-7 | 0.511 442 | 2.655 830 | 0.192 573 |
| VvSKP1-10 | VvSKP1-6 | 0.403 211 | 2.906 73 | 0.138 717 | VvSKP1-5 | VvSKP1-6 | 0.380 355 | 3.273 070 | 0.116 207 |
| VvSKP1-10 | VvSKP1-7 | 0.545 177 | 2.499 55 | 0.218 110 | VvSKP1-5 | VvSKP1-7 | 0.273 146 | 3.618 320 | 0.075 489 9 |
| VvSKP1-11 | VvSKP1-3 | 0.718 013 | 1.848 36 | 0.388 460 | VvSKP1-6 | VvSKP1-7 | 0.516 509 | 2.610 210 | 0.197 880 |
表3 葡萄SKP1基因家族共线性关系及非同义替换率(Ka)和同义替换率(Ks)
Table 3 Collinearity relationships of grape SKP1 gene family and non-synonymous substitution rate (Ka) and synonymous substitution rate (Ks)
共线性序列1 Collinearity sequence 1 | 共线性序列2 Collinearity sequence 2 | Ka | Ks | Ka/Ks | 共线性序列1 Collinearity sequence 1 | 共线性序列2 Collinearity sequence 2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|---|---|---|---|---|
| VvSKP1-1 | VvSKP1-10 | 0.484 186 | 2.676 18 | 0.180 925 | VvSKP1-11 | VvSKP1-4 | 0.318 353 | 0.978 672 | 0.325 291 |
| VvSKP1-1 | VvSKP1-11 | 0.439 432 | 2.748 94 | 0.159 855 | VvSKP1-11 | VvSKP1-5 | 0.394 356 | 3.214 360 | 0.122 685 |
| VvSKP1-1 | VvSKP1-3 | 0.744 929 | 1.804 00 | 0.412 931 | VvSKP1-11 | VvSKP1-6 | 0.312 299 | 0.961 286 | 0.324 876 |
| VvSKP1-1 | VvSKP1-4 | 0.374 420 | 3.230 87 | 0.115 888 | VvSKP1-11 | VvSKP1-7 | 0.559 368 | 2.658 880 | 0.210 377 |
| VvSKP1-1 | VvSKP1-5 | 0.401 063 | 3.083 55 | 0.130 065 | VvSKP1-3 | VvSKP1-4 | 0.657 090 | 2.075 610 | 0.316 577 |
| VvSKP1-1 | VvSKP1-6 | 0.378 107 | 3.186 02 | 0.118 677 | VvSKP1-3 | VvSKP1-5 | 0.526 860 | 2.452 040 | 0.214 866 |
| VvSKP1-1 | VvSKP1-7 | 0.551 033 | 2.400 07 | 0.229 591 | VvSKP1-3 | VvSKP1-6 | 0.674 153 | 2.036 490 | 0.331 038 |
| VvSKP1-10 | VvSKP1-11 | 0.473 489 | 2.443 63 | 0.193 765 | VvSKP1-3 | VvSKP1-7 | 0.810 727 | 1.581 060 | 0.512 774 |
| VvSKP1-10 | VvSKP1-3 | 0.639 464 | 2.206 82 | 0.289 767 | VvSKP1-4 | VvSKP1-5 | 0.364 465 | 3.387 940 | 0.107 577 |
| VvSKP1-10 | VvSKP1-4 | 0.402 901 | 2.944 32 | 0.136 840 | VvSKP1-4 | VvSKP1-6 | 0.010 264 2 | 0.014 291 7 | 0.718 192 |
| VvSKP1-10 | VvSKP1-5 | 0.384 626 | 3.164 50 | 0.121 544 | VvSKP1-4 | VvSKP1-7 | 0.511 442 | 2.655 830 | 0.192 573 |
| VvSKP1-10 | VvSKP1-6 | 0.403 211 | 2.906 73 | 0.138 717 | VvSKP1-5 | VvSKP1-6 | 0.380 355 | 3.273 070 | 0.116 207 |
| VvSKP1-10 | VvSKP1-7 | 0.545 177 | 2.499 55 | 0.218 110 | VvSKP1-5 | VvSKP1-7 | 0.273 146 | 3.618 320 | 0.075 489 9 |
| VvSKP1-11 | VvSKP1-3 | 0.718 013 | 1.848 36 | 0.388 460 | VvSKP1-6 | VvSKP1-7 | 0.516 509 | 2.610 210 | 0.197 880 |

图6 葡萄SKP1基因家族成员接种灰霉病后的表达水平不同字母表示差异显著(P<0.05),下同
Fig. 6 Expressions of grape SKP1 gene family members after inoculation with Botrytis cinereaThe different letters indicate significant differences (P<0.05). The same below
| [1] | Sharafan M, Malinowska MA, Ekiert H, et al. Vitis vinifera (vine grape) as a valuable cosmetic raw material [J]. Pharmaceutics, 2023, 15(5): 1372. |
| [2] | 郭泽西, 孙大运, 曲俊杰, 等. 查尔酮合成酶基因在葡萄抗灰霉病和霜霉病中的作用 [J]. 中国农业科学, 2022, 55(6): 1139-1148. |
| Guo ZX, Sun DY, Qu JJ, et al. The role of chalcone synthase gene in grape resistance to gray mold and downy mildew [J]. Sci Agric Sin, 2022, 55(6): 1139-1148. | |
| [3] | He F, Niu MX, Wang T, et al. The ubiquitin E3 ligase RZFP1 affects drought tolerance in poplar by mediating the degradation of the protein phosphatase PP2C-9 [J]. Plant Physiol, 2024, 196(4): 2936-2955. |
| [4] | Xu J, Liu HJ, Zhou C, et al. The ubiquitin-proteasome system in the plant response to abiotic stress: Potential role in crop resilience improvement [J]. Plant Sci, 2024, 342: 112035. |
| [5] | Majumdar P, Karidas P, Siddiqi I, et al. The ubiquitin-specific protease TNI/UBP14 functions in ubiquitin recycling and affects auxin response [J]. Plant Physiol, 2020, 184(3): 1499-1513. |
| [6] | Ali SM, Li N, Soufi Z, et al. Multiple ubiquitin E3 ligase genes antagonistically regulate chloroplast-associated protein degradation [J]. Curr Biol, 2023, 33(6): 1138-1146.e5. |
| [7] | Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology [J]. Nat Rev Mol Cell Biol, 2009, 10(6): 385-397. |
| [8] | Zhou Y, Huang YD, Wu L, et al. Phylogenetic and expression analyses of cullin family members unveil the role of PbCUL1.C1 in pollen tube growth underlying non-self S-RNase in pear [J]. Plant Mol Biol Report, 2020, 38(4): 601-612. |
| [9] | Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway [J]. Annu Rev Plant Biol, 2004, 55: 555-590. |
| [10] | Nayak S, Santiago FE, Jin H, et al. The Caenorhabditis elegans Skp1-related gene family diverse functions in cell proliferation, morphogenesis, and meiosis [J]. Curr Biol, 2002, 12(4): 277-287. |
| [11] | Xu LH, Liu FQ, Lechner E, et al. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis [J]. Plant Cell, 2002, 14(8): 1919-1935. |
| [12] | Ni WM, Xie DX, Hobbie L, et al. Regulation of flower development in Arabidopsis by SCF complexes [J]. Plant Physiol, 2004, 134(4): 1574-1585. |
| [13] | Kong HZ, Leebens-Mack J, Ni WM, et al. Highly heterogeneous rates of evolution in the SKP1 gene family in plants and animals: functional and evolutionary implications [J]. Mol Biol Evol, 2004, 21(1): 117-128. |
| [14] | Dezfulian MH, Soulliere DM, Dhaliwal RK, et al. The SKP1-like gene family of Arabidopsis exhibits a high degree of differential gene expression and gene product interaction during development [J]. PLoS One, 2012, 7(11): e50984. |
| [15] | Kahloul S, HajSalah El Beji I, Boulaflous A, et al. Structural, expression and interaction analysis of rice SKP1-like genes [J]. DNA Res, 2013, 20(1): 67-78. |
| [16] | Li YZ, Lin W, Zhu JW, et al. Genome-wide analysis of the S-phase kinase-association protein 1 (ClSKP1) family and the role of S-RNase targeting by an SCF (Cullin1-SKP1-F-box) complex in the self-incompatibility of ‘Xiangshui’ lemon [J]. Hortic Plant J, 2025, 11(2): 593-607. |
| [17] | Shao M, Wang P, Gou HM, et al. Identification and expression analysis of the SKP1-like gene family under phytohormone and abiotic stresses in apple (Malus domestica) [J]. Int J Mol Sci, 2023, 24(22): 16414. |
| [18] | Varshney V, Hazra A, Majee M. Identification, genomic organization, and comprehensive expression analysis reveals the implication of Cicer arietinum SKP1-like genes in abiotic stress [J]. J Plant Growth Regul, 2023, 42(10): 6074-6090. |
| [19] | 张春渝, 许小琼, 徐小萍, 等. 龙眼SKP1-like家族成员鉴定及体胚发生早期表达分析 [J]. 园艺学报, 2021, 48(9): 1665-1679. |
| Zhang CY, Xu XQ, Xu XP, et al. Genome-wide identification of the SKP1-like family and analysis of their expression during early somatic embryogenesis in Longan [J]. Acta Hortic Sin, 2021, 48(9): 1665-1679. | |
| [20] | Duyvesteijn RGE, Van Wijk R, Boer Y, et al. Frp1 is a Fusarium oxysporum F-box protein required for pathogenicity on tomato [J]. Mol Microbiol, 2005, 57(4): 1051-1063. |
| [21] | Alkharouf NW, Klink VP, Chouikha IB, et al. Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode) [J]. Planta, 2006, 224(4): 838-852. |
| [22] | Ning B, Li WW, Liu X, et al. GmSKP1, a novel S-phase kinase-associated protein 1 in Glycine max, enhancing resistance against Phytophthora sojae infection [J]. J Northeast Agric Univ Engl Ed, 2023, 30(1): 1-12. |
| [23] | Ban ZN, Estelle M. CUL3 E3 ligases in plant development and environmental response [J]. Nat Plants, 2021, 7(1): 6-16. |
| [24] | Sharma S, Prasad A, Prasad M. Ubiquitination from the perspective of plant pathogens [J]. J Exp Bot, 2023, 74(15): 4367-4376. |
| [25] | Mistry J, Finn RD, Eddy SR, et al. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions [J]. Nucleic Acids Res, 2013, 41(12): e121. |
| [26] | Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool [J]. J Mol Biol, 1990, 215(3): 403-410. |
| [27] | Bateman A, Birney E, Durbin R, et al. The Pfam protein families database [J]. Nucleic Acids Res, 2000, 28(1): 263-266. |
| [28] | Finn RD, Bateman A, Clements J, et al. Pfam: the protein families database [J]. Nucleic Acids Res, 2014, 42(Database issue): D222-D230. |
| [29] | Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal [J]. Nucleic Acids Res, 2012, 40(Web Server issue): W597-W603. |
| [30] | Horton P, Park KJ, Obayashi T, et al. WoLF PSORT: protein localization predictor [J]. Nucleic Acids Res, 2007, 35(Web Server issue): W585-W587. |
| [31] | Petersen TN, Brunak S, von Heijne G, et al. SignalP 4.0: discriminating signal peptides from transmembrane regions [J]. Nat Methods, 2011, 8(10): 785-786. |
| [32] | Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability [J]. Mol Biol Evol, 2013, 30(4): 772-780. |
| [33] | Kumar S, Nei M, Dudley J, et al. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences [J]. Brief Bioinform, 2008, 9(4): 299-306. |
| [34] | Waterhouse AM, Procter JB, Martin DMA, et al. Jalview Version 2—a multiple sequence alignment editor and analysis workbench [J]. Bioinformatics, 2009, 25(9): 1189-1191. |
| [35] | Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server [J]. Bioinformatics, 2015, 31(8): 1296-1297. |
| [36] | Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching [J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| [37] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| [38] | Chao JT, Li ZY, Sun YH, et al. MG2C: a user-friendly online tool for drawing genetic maps [J]. Mol Hortic, 2021, 1(1): 16. |
| [39] | Wang YP, Tang HB, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity [J]. Nucleic Acids Res, 2012, 40(7): e49. |
| [40] | Zhang Z, Li J, Zhao XQ, et al. KaKs_Calculator: calculating ka and ks through model selection and model averaging [J]. Genom Proteom Bioinform, 2006, 4(4): 259-263. |
| [41] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔct Method [J]. Methods, 2001, 25(4): 402-408. |
| [42] | Huang SX, Sirikhachornkit A, Su XJ, et al. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat [J]. Proc Natl Acad Sci USA, 2002, 99(12): 8133-8138. |
| [43] | Jeffares DC, Penkett CJ, Bähler J. Rapidly regulated genes are intron poor [J]. Trends Genet, 2008, 24(8): 375-378. |
| [44] | 范润侨, 魏春茹, 杨一鸣, 等. 小麦SKP1家族成员鉴定、基因表达及其与F-box蛋白的互作分析 [J]. 农业生物技术学报, 2022, 30(2): 207-221. |
| Fan RQ, Wei CR, Yang YM, et al. Identification and expression analysis of wheat (Triticum aestivum) SKP1 family members and their interaction with F-box proteins [J]. J Agric Biotechnol, 2022, 30(2): 207-221. | |
| [45] | 曹华盛, 李堂, 熊亮, 等. 水稻溶血磷脂酸酰基转移酶基因(LPAT)家族生物信息学分析及在籽粒油脂合成中的作用 [J]. 华南农业大学学报, 2023, 44(6): 925-935. |
| Cao HS, Li T, Xiong L, et al. Bioinformatics analysis of rice lysophosphatidic acid acyltransferase gene (LPAT) family and its function in the oil synthesis of rice grains [J]. J South China Agric Univ, 2023, 44(6): 925-935. | |
| [46] | Wang Q, Tao T, Han YH, et al. Nonstructural protein P7-2 encoded by Rice black-streaked dwarf virus interacts with SKP1, a core subunit of SCF ubiquitin ligase [J]. Virol J, 2013, 10: 325. |
| [47] | Liu YL, Schiff M, Serino G, et al. Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus [J]. Plant Cell, 2002, 14(7): 1483-1496. |
| [48] | Su YY, Ngea GLN, Wang KL, et al. Deciphering the mechanism of E3 ubiquitin ligases in plant responses to abiotic and biotic stresses and perspectives on PROTACs for crop resistance [J]. Plant Biotechnol J, 2024, 22(10): 2811-2843. |
| [49] | Li XC, Sun Y, Liu NN, et al. Enhanced resistance to Verticillium dahliae mediated by an F-box protein GhACIF1 from Gossypium hirsutum [J]. Plant Sci, 2019, 284: 127-134. |
| [50] | 吴渊源, 韦鹏飞, 郭崇炎, 等. 小粒野生稻OmSKP1基因的克隆与功能初探 [J]. 分子植物育种, 2019, 17(23): 7641-7648. |
| Wu YY, Wei PF, Guo CY, et al. Cloning and functional analysis of OmSKP1 gene from Oryza minuta [J]. Mol Plant Breed, 2019, 17(23): 7641-7648. |
| [1] | 董向向, 缪百灵, 许贺娟, 陈娟娟, 李亮杰, 龚守富, 朱庆松. 森林草莓FveBBX20基因的生物信息学分析及开花调控功能[J]. 生物技术通报, 2025, 41(9): 115-123. |
| [2] | 化文平, 刘菲, 浩佳欣, 陈尘. 丹参ADH基因家族的鉴定与表达模式分析[J]. 生物技术通报, 2025, 41(8): 211-219. |
| [3] | 腊贵晓, 赵玉龙, 代丹丹, 余永亮, 郭红霞, 史贵霞, 贾慧, 杨铁钢. 红花质膜H+-ATPase基因家族成员鉴定及响应低氮低磷胁迫的表达分析[J]. 生物技术通报, 2025, 41(8): 220-233. |
| [4] | 赖诗雨, 梁巧兰, 魏列新, 牛二波, 陈应娥, 周鑫, 杨思正, 王博. NbJAZ3在苜蓿花叶病毒侵染本氏烟过程中的作用[J]. 生物技术通报, 2025, 41(8): 186-196. |
| [5] | 任睿斌, 司二静, 万广有, 汪军成, 姚立蓉, 张宏, 马小乐, 李葆春, 王化俊, 孟亚雄. 大麦条纹病菌GH17基因家族的鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 146-154. |
| [6] | 曾丹, 黄园, 王健, 张艳, 刘庆霞, 谷荣辉, 孙庆文, 陈宏宇. 铁皮石斛bZIP转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 197-210. |
| [7] | 程雪, 付颖, 柴晓娇, 王红艳, 邓欣. 谷子LHC基因家族鉴定及非生物胁迫表达分析[J]. 生物技术通报, 2025, 41(8): 102-114. |
| [8] | 白雨果, 李婉迪, 梁建萍, 石志勇, 卢庚龙, 刘红军, 牛景萍. 哈茨木霉T9131对黄芪幼苗的促生机理[J]. 生物技术通报, 2025, 41(8): 175-185. |
| [9] | 侯鹰翔, 费思恬, 黎妮, 李兰, 宋松泉, 王伟平, 张超. 水稻miRNAs响应生物胁迫研究进展[J]. 生物技术通报, 2025, 41(7): 69-80. |
| [10] | 李凯月, 邓晓霞, 殷缘, 杜亚彤, 徐元静, 王竞红, 于耸, 蔺吉祥. 蓖麻LEA基因家族的鉴定和铝胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 128-138. |
| [11] | 牛景萍, 赵婧, 郭茜, 王书宏, 赵晋忠, 杜维俊, 殷丛丛, 岳爱琴. 基于WGCNA鉴定大豆抗大豆花叶病毒NAC转录因子及其诱导表达分析[J]. 生物技术通报, 2025, 41(7): 95-105. |
| [12] | 韩燚, 侯昌林, 唐露, 孙璐, 谢晓东, 梁晨, 陈小强. 大麦HvERECTA基因的克隆及功能分析[J]. 生物技术通报, 2025, 41(7): 106-116. |
| [13] | 李霞, 张泽伟, 刘泽军, 王楠, 郭江波, 辛翠花, 张彤, 简磊. 马铃薯转录因子StMYB96的克隆及功能研究[J]. 生物技术通报, 2025, 41(7): 181-192. |
| [14] | 龚钰涵, 陈兰, 尚方慧子, 郝灵颖, 刘硕谦. 茶树TRB基因家族鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(7): 214-225. |
| [15] | 魏雨佳, 李岩, 康语涵, 弓晓楠, 杜敏, 涂岚, 石鹏, 于子涵, 孙彦, 张昆. 白颖苔草CrMYB4基因的克隆和表达分析[J]. 生物技术通报, 2025, 41(7): 248-260. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||