








生物技术通报 ›› 2026, Vol. 42 ›› Issue (1): 76-85.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0975
任泓宇1,2( ), 庞翠萍1, 古阳1(
), 庞翠萍1, 古阳1( ), 周佳海1,3(
), 周佳海1,3( )
)
收稿日期:2025-09-11
出版日期:2026-01-26
发布日期:2026-02-04
通讯作者:
周佳海,男,博士,教授,研究方向 :结构生物学、天然产物生物合成;E-mail: jiahai@siat.ac.cn作者简介:任泓宇,男,硕士研究生,研究方向 :结构生物学;E-mail: hy.ren@siat.ac.cn
基金资助:
REN Hong-yu1,2( ), PANG Cui-ping1, GU Yang1(
), PANG Cui-ping1, GU Yang1( ), ZHOU Jia-hai1,3(
), ZHOU Jia-hai1,3( )
)
Received:2025-09-11
Published:2026-01-26
Online:2026-02-04
摘要:
目的 蛋白质晶体衍射是结构生物学研究的重要方法,晶体的衍射分辨率直接决定了模型的精确度及应用可行性。需要采取多种策略组合优化蛋白质晶体质量,提高晶体的衍射分辨率。 方法 以催化芳基偶联反应的P450酶为研究对象,利用分子生物学技术构建原核表达系统,并通过镍柱亲和层析和体积排阻色谱等方法进行纯化。在结晶阶段,采用气相扩散法系统筛选了1 632种初始条件,以此为基础,重点对缓冲液pH值、沉淀剂种类与浓度等关键变量进行精细化筛选,并引入多种添加剂以改善结晶环境。进一步结合结构预测分析,对蛋白柔性区域进行理性截短设计,并尝试SUMO标签的融合表达以增强蛋白的稳定性。 结果 实现了P450蛋白在大肠杆菌中的高效可溶表达,并经两步纯化获得可用于结晶的高纯度蛋白。通过大规模结晶条件筛选与多轮优化,结合序列截短和SUMO标签融合等策略协同作用,晶体形态显著改善,由原来的微晶或无定形沉淀转变为外观规则、棱角清晰的单晶。晶体衍射分辨率由初始的10 Å显著提升至2.86 Å,获得了可用于高分辨率结构解析的优质晶体。 结论 使用多策略协同的方法成功改善了蛋白晶体形态,提高了蛋白晶体的衍射分辨率。
任泓宇, 庞翠萍, 古阳, 周佳海. 多策略协同提升蛋白质晶体的衍射分辨率[J]. 生物技术通报, 2026, 42(1): 76-85.
REN Hong-yu, PANG Cui-ping, GU Yang, ZHOU Jia-hai. Multi-strategy Synergy Enhances the Diffraction Resolution of Protein Crystals[J]. Biotechnology Bulletin, 2026, 42(1): 76-85.

图1 P450表达标签替换及分子伴侣共表达A-C:分别为P450KstB、P450MciB、P450ScnB添加分子伴侣及替换融合标签的表达情况,其中编号1为未诱导菌株,编号2为SUMO-P450,编号3-5为SUMO-P450分别添加分子伴侣pGro7、pTF16和pKJE7,编号6为TF-P450,编号7为MBP-P450;D:替换GST标签的表达情况,编号1为未诱导菌株,编号2-4分别为添加GST标签的P450KstB、P450ScnB和P450MciB
Fig. 1 Replacing expression tags and co-expressing molecular chaperones of P450A-C: The expression of P450KstB, P450MciB, and P450ScnB, respectively, with molecular chaperone co-expression and fusion tag swapping. 1: Uninduced strain, 2: SUMO-P450, 3-5: the SUMO-P450 strain was introduced with the molecular chaperones pGro7, pTF16, and pKJE7, respectively, 6: TF-P450, 7: MBP-P450. D: The expression after replacing the GST tag. 1: Uninduced strain, 2-4: P450KstB, P450ScnB, and P450MciB fused with GST tag, respectively
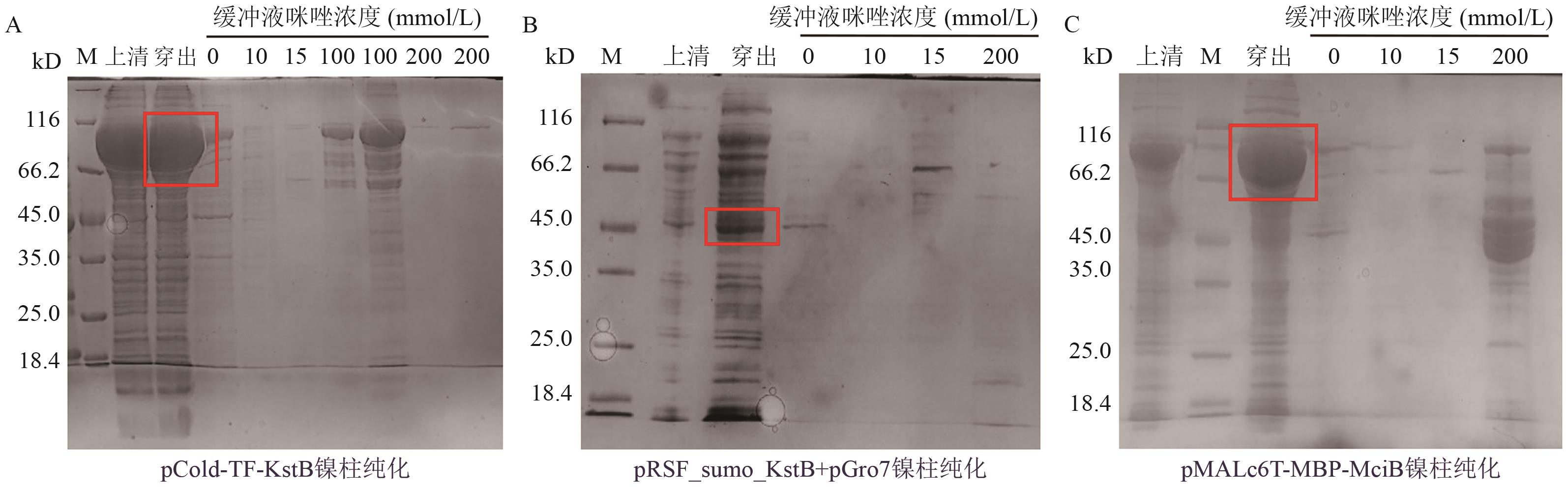
图2 可溶蛋白的Ni柱亲和层析结果分析A-C:分别为使用镍柱对pCold-TF-KstB、pRSF-sumo-KstB+pGro7和pMALc6T-MBP-MciB进行亲和层析的纯化结果
Fig. 2 Ni-NTA analysis of soluble proteins results by Ni-affinity chromatographyA-C: The purification results of affinity chromatography using nickel columns for pCold-TF-KstB, pRSF-sumo-KstB+pGro7, and pMALc6T-MBP-MciB, respectively

图3 P450GpeC蛋白的表达情况分析A:P450GpeC的细胞破碎液及沉淀/上清的SDS-PAGE分析;B:P450GpeC的紫外吸收光谱,425 nm处为特征吸收
Fig. 3 Analysis of P450GpeC protein expressionA: The cell lysate of P450GpeC and the SDS-PAGE analysis of precipitate and supernatant. B: The UV absorption spectrum of P450GpeC with a characteristic absorption peak at 425 nm
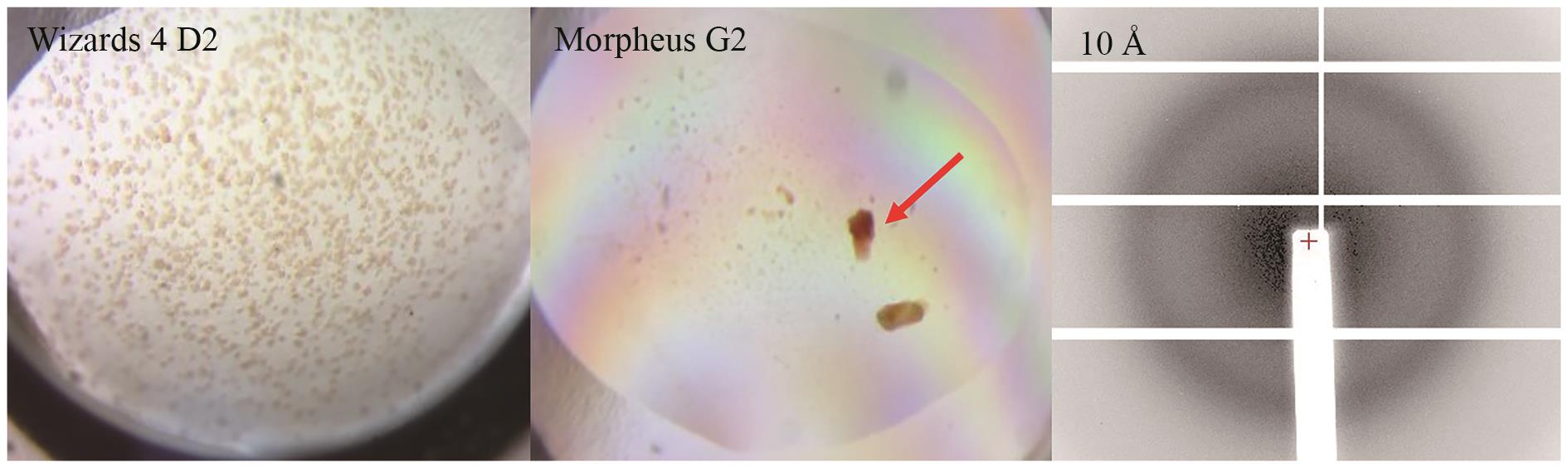
图7 P450GpeC初筛后不同结晶条件下的晶体形态及衍射图箭头所示为用于衍射的晶体
Fig. 7 Crystal images and diffraction patterns of P450GpeC under various initial crystallization conditionsThe arrow indicates the crystal used for diffraction
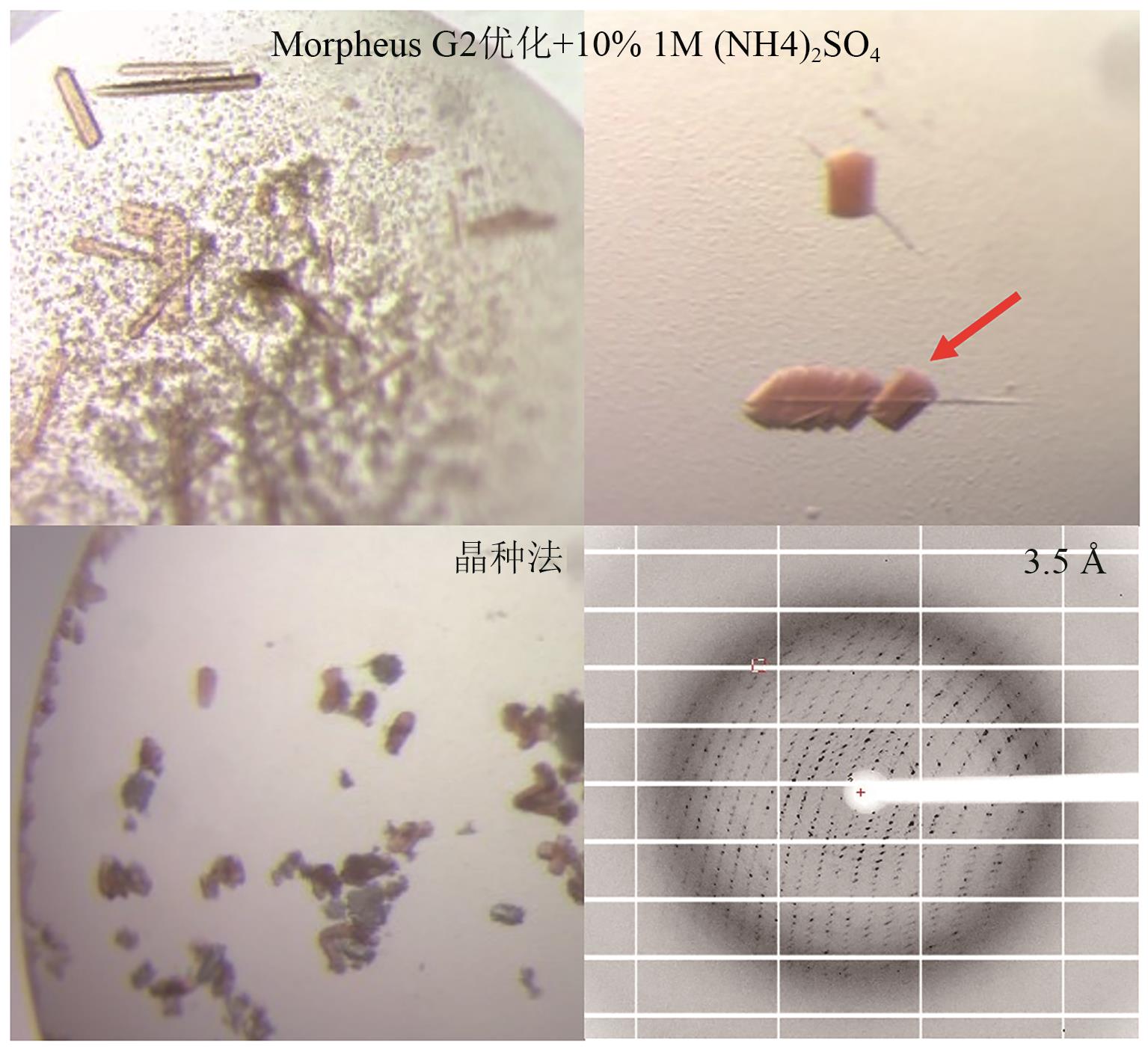
图8 添加硫酸铵及晶种法优化后的P450GpeC晶体及衍射图箭头所示为用于衍射的晶体
Fig. 8 Crystal images and diffraction patterns of P450GpeC optimized by ammonium sulfate addition and seeding methodThe arrow indicates the crystal used for diffraction

图11 对SUMO-P450GpeC-△C8优化后的晶体形态及衍射图箭头所示为用于衍射的晶体
Fig. 11 Crystal images and diffraction patterns of optimized SUMO-P450GpeC-△C8 crystalsThe arrow indicates the crystal used for diffraction
优化方式 Optimization method | 优化效果 Optimization result | 分辨率 Resolution (Å) |
|---|---|---|
| 结晶初筛 | 无法获得蛋白晶体 | - |
| C端loop截短 | 晶体过小或者表面粗糙 | 10 |
| 溶剂优化 | 改善晶体形态 | 5-8 |
| 添加硫酸铵 | 依附盐晶生长 | 3-4 |
| 添加晶种 | 脱离盐晶 | 3-4 |
| 添加SUMO标签 | 外观规则且棱角清晰的高质量集体 | 2.86 |
表1 晶体优化方式及效果
Table 1 Crystal optimization methods and effects
优化方式 Optimization method | 优化效果 Optimization result | 分辨率 Resolution (Å) |
|---|---|---|
| 结晶初筛 | 无法获得蛋白晶体 | - |
| C端loop截短 | 晶体过小或者表面粗糙 | 10 |
| 溶剂优化 | 改善晶体形态 | 5-8 |
| 添加硫酸铵 | 依附盐晶生长 | 3-4 |
| 添加晶种 | 脱离盐晶 | 3-4 |
| 添加SUMO标签 | 外观规则且棱角清晰的高质量集体 | 2.86 |
| [1] | Papageorgiou AC, Poudel N, Mattsson J. Protein structure analysis and validation with X-ray crystallography [M]//Protein Downstream Processing. New York, NY: Springer US, 2020: 377-404. |
| [2] | Banari A, Samanta AK, Munke A, et al. Advancing time-resolved structural biology: latest strategies in cryo-EM and X-ray crystallography [J]. Nat Meth, 2025, 22(7): 1420-1435. |
| [3] | Nogales E, Mahamid J. Bridging structural and cell biology with cryo-electron microscopy [J]. Nature, 2024, 628(8006): 47-56. |
| [4] | Abramson J, Adler J, Dunger J, et al. Addendum: Accurate structure prediction of biomolecular interactions with AlphaFold 3 [J]. Nature, 2024, 636(8042): E4. |
| [5] | Li FC, Liu RZ, Li WJ, et al. Synchrotron radiation: a key tool for drug discovery [J]. Bioorg Med Chem Lett, 2024, 114: 129990. |
| [6] | Heras B, Edeling MA, Byriel KA, et al. Dehydration converts DsbG crystal diffraction from low to high resolution [J]. Structure, 2003, 11(2): 139-145. |
| [7] | McPherson A, Gavira JA. Introduction to protein crystallization [J]. Acta Crystallogr F Struct Biol Commun, 2014, 70(1): 2-20. |
| [8] | Budziszewski GR, Stojanoff V, Bowman SEJ. Preparing for successful protein crystallization experiments [J]. Acta Crystallogr F Struct Biol Commun, 2025, 81(7): 272-280. |
| [9] | Zhang XF, Xu ZT, Zhou JH, et al. Enhancement of protein crystallization using nano-sized metal-organic framework [J]. Crystals, 2022, 12(5): 578. |
| [10] | Maki S, Hagiwara M. Contactless crystallization method of protein by a magnetic force booster [J]. Sci Rep, 2022, 12: 17287. |
| [11] | Han Q, Brown SJ, Drummond CJ, et al. Protein aggregation and crystallization with ionic liquids: Insights into the influence of solvent properties [J]. J Colloid Interface Sci, 2022, 608: 1173-1190. |
| [12] | Chen TE, Hiramatsu H, Toyouchi S, et al. Laser-polarization-induced anisotropy enhances protein crystallization [J]. Angew Chem Int Ed, 2025, 64(21): e202501827. |
| [13] | Dessau MA, Modis Y. Protein crystallization for X-ray crystallography [J]. JoVE, 2011(47). |
| [14] | Chayen NE, Saridakis E. Protein crystallization: from purified protein to diffraction-quality crystal [J]. Nat Meth, 2008, 5(2): 147-153. |
| [15] | McPherson A, Cudney B. Searching for silver bullets: an alternative strategy for crystallizing macromolecules [J]. J Struct Biol, 2006, 156(3): 387-406. |
| [16] | de Wijn R, Hennig O, Ernst FGM, et al. Combining crystallogenesis methods to produce diffraction-quality crystals of a psychrophilic tRNA-maturation enzyme [J]. Acta Crystallogr F Struct Biol Commun, 2018, 74(11): 747-753. |
| [17] | Khurshid S, Saridakis E, Govada L, et al. Porous nucleating agents for protein crystallization [J]. Nat Protoc, 2014, 9(7): 1621-1633. |
| [18] | Chernov AA. Protein crystals and their growth [J]. J Struct Biol, 2003, 142(1): 3-21. |
| [19] | Zhao MZ, Ma JS, Li M, et al. Cytochrome P450 enzymes and drug metabolism in humans [J]. Int J Mol Sci, 2021, 22(23): 12808. |
| [20] | Sang RY, Jiang WZ, Zhang C, et al. Effect of food components on cytochrome P450 expression and activity [J]. Hum Nutr Metab, 2025, 40: 200304. |
| [21] | Song YR, Li CX, Liu GZ, et al. Drug-metabolizing cytochrome P450 enzymes have multifarious influences on treatment outcomes [J]. Clin Pharmacokinet, 2021, 60(5): 585-601. |
| [22] | Esteves F, Rueff J, Kranendonk M. The central role of cytochrome P450 in xenobiotic metabolism—a brief review on a fascinating enzyme family [J]. J Xenobiot, 2021, 11(3): 94-114. |
| [23] | Greule A, Stok JE, De Voss JJ, et al. Unrivalled diversity: the many roles and reactions of bacterial cytochromes P450 in secondary metabolism 1 [J]. Nat Prod Rep, 2018, 35(8): 757-791. |
| [24] | Rudolf JD, Chang CY, Ma M, et al. Cytochromes P450 for natural product biosynthesis in Streptomyces: sequence, structure, and function [J]. Nat Prod Rep, 2017, 34(9): 1141-1172. |
| [25] | Zhang XW, Guo JW, Cheng FY, et al. Cytochrome P450 enzymes in fungal natural product biosynthesis [J]. Nat Prod Rep, 2021, 38(6): 1072-1099. |
| [26] | He BB, Liu J, Cheng Z, et al. Bacterial cytochrome P450 catalyzed post-translational macrocyclization of ribosomal peptides [J]. Angew Chem Int Ed, 2023, 62(46): e202311533. |
| [27] | Hu YL, Yin FZ, Shi J, et al. P450-modified ribosomally synthesized peptides with aromatic cross-links [J]. J Am Chem Soc, 2023, 145(50): 27325-27335. |
| [28] | Coe LJ, Zhao YW, Padva L, et al. Reassignment of the structure of a tryptophan-containing cyclic tripeptide produced by the biarylitide crosslinking cytochrome P450blt [J]. Chem, 2024, 30(38): e202400988. |
| [29] | Kandy SK, Pasquale MA, Chekan JR. Aromatic side-chain crosslinking in RiPP biosynthesis [J]. Nat Chem Biol, 2025, 21(2): 168-181. |
| [30] | Zdouc MM, Alanjary MM, Zarazúa GS, et al. A biaryl-linked tripeptide from Planomonospora reveals a widespread class of minimal RiPP gene clusters [J]. Cell Chem Biol, 2021, 28(5): 733-739.e4. |
| [31] | Hug JJ, Dastbaz J, Adam S, et al. Biosynthesis of cittilins, unusual ribosomally synthesized and post-translationally modified peptides from Myxococcus xanthus [J]. ACS Chem Biol, 2020, 15(8): 2221-2231. |
| [32] | Wang F, Zhou HY, Olademehin OP, et al. Insights into key interactions between vancomycin and bacterial cell wall structures [J]. ACS Omega, 2018, 3(1): 37-45. |
| [33] | Hu BD, Yu HB, Zhou JW, et al. Whole-cell P450 biocatalysis using engineered Escherichia coli with fine-tuned heme biosynthesis [J]. Adv Sci, 2023, 10(6): 2205580. |
| [34] | Panavas T, Sanders C, Butt TR. SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems [J]. Methods Mol Biol, 2009, 497: 303-317. |
| [1] | 王嘉, 高暝, 赵耘霄, 陈益存, 汪阳东. 细胞色素P450参与次生代谢物合成响应生物胁迫[J]. 生物技术通报, 2025, 41(12): 27-39. |
| [2] | 谭景轩, 邢德勋, 何天锦, 刘占英. 荧光假单胞菌蛋白表达系统研究进展[J]. 生物技术通报, 2025, 41(1): 49-61. |
| [3] | 潘萍萍, 徐志浩, 张怡雯, 李青, 王忠华. 多花黄精查尔酮合酶PcCHS的原核表达、亚细胞定位及表达分析[J]. 生物技术通报, 2024, 40(5): 280-289. |
| [4] | 张岩峰, 叶丽丹, 于洪巍. 氧化还原伴侣工程:P450低效问题的解决方案之一[J]. 生物技术通报, 2023, 39(4): 10-23. |
| [5] | 郁慧丽, 李爱涛. 细胞色素P450酶在香精香料绿色生物合成中的应用[J]. 生物技术通报, 2023, 39(4): 24-37. |
| [6] | 覃雪晶, 王雨涵, 曹一博, 张凌云. 青杄PwHAP5基因原核表达及多克隆抗体制备[J]. 生物技术通报, 2022, 38(8): 142-149. |
| [7] | 魏倩, 刘小宁, 赵洁. 2-十三烷酮胁迫下棉铃虫FoxAl调控CYP6B6的表达[J]. 生物技术通报, 2022, 38(5): 84-92. |
| [8] | 杨佳慧, 孙玉萍, 陆雅宁, 刘欢, 卢存福, 陈玉珍. 拟南芥AtTERT对大肠杆菌非生物胁迫抗性的影响[J]. 生物技术通报, 2022, 38(2): 1-9. |
| [9] | 曹汝菲, 李泽轩, 许欢, 张莎, 张敏敏, 戴枫, 段晓雷. 脆弱拟杆菌Pif1解旋酶的表达纯化与晶体生长[J]. 生物技术通报, 2021, 37(9): 180-190. |
| [10] | 范晨龙, 丁燏. 溶藻弧菌去乙酰化酶基因cobB克隆及其功能验证[J]. 生物技术通报, 2021, 37(8): 195-202. |
| [11] | 唐禄, 董丽平, 尹茉莉, 刘磊, 董媛, 王会岩. 成纤维细胞生长因子20单克隆抗体的制备及鉴定[J]. 生物技术通报, 2021, 37(10): 179-185. |
| [12] | 杨世全, 彭丹, 费文杰, 杨丰, 屈高毅, 唐威威, 欧剑萍, 邓湘雯, 周波. 杉木ClKptA/Tpt1基因的克隆及其表达特性分析[J]. 生物技术通报, 2020, 36(8): 15-22. |
| [13] | 闵琪, 高子涵, 姚银, 张华山, 熊海容, 张莉. 共表达HAC1和分子伴侣基因对甘露聚糖酶在毕赤酵母中表达的影响[J]. 生物技术通报, 2020, 36(5): 159-168. |
| [14] | 孟利, 杜彩萍. 大鼠His-Akt1重组蛋白的真核表达、蛋白纯化及活性鉴定[J]. 生物技术通报, 2020, 36(12): 98-103. |
| [15] | 周阳, 王桃桃, 闫丹丹, 王莹莹, 施沁璇, 孙丽慧, 林锋. 类弹性蛋白作为功能纳米材料在生物工程领域研究及应用进展[J]. 生物技术通报, 2020, 36(11): 198-208. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||