








生物技术通报 ›› 2026, Vol. 42 ›› Issue (1): 86-94.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0542
杨跃琴1( ), 邢英1(
), 邢英1( ), 仲子荷2,3, 田维军3,4, 杨雪清3,4, 王建旭3
), 仲子荷2,3, 田维军3,4, 杨雪清3,4, 王建旭3
收稿日期:2025-05-26
出版日期:2026-01-26
发布日期:2026-02-04
通讯作者:
邢英,女,博士,教授,研究方向 :重金属土壤环境化学;E-mail: xy31034@163.com作者简介:杨跃琴,女,硕士研究生,研究方向 :环境污染化学;E-mail: yangyueqin3569@163.com
基金资助:
YANG Yue-qin1( ), XING Ying1(
), XING Ying1( ), ZHONG Zi-he2,3, TIAN Wei-jun3,4, YANG Xue-qing3,4, WANG Jian-xu3
), ZHONG Zi-he2,3, TIAN Wei-jun3,4, YANG Xue-qing3,4, WANG Jian-xu3
Received:2025-05-26
Published:2026-01-26
Online:2026-02-04
摘要:
目的 多药和有毒化合物外排转运蛋白(MATE)在植物营养物质吸收、次生代谢物转运、重金属和外源物质解毒等方面发挥重要的调控作用。研究水稻OsMATE34的生物学功能,为深入理解水稻富集MeHg的分子机制奠定基础,亦为培育低MeHg水稻品种提供科学参考。 方法 通过生物信息学方法,分析OsMATE34的理化性质和系统进化树,利用实时荧光定量PCR(RT-qPCR)检测MeHg胁迫下水稻不同组织OsMATE34的相对表达量,并研究MeHg胁迫后水稻不同组织MeHg含量,用以分析其表达量与MeHg含量之间的相关性。利用CRISPR/Cas9基因编辑技术,获得1个OsMATE34敲除株系,在苗期进行MeHg处理,测定野生型(WT)和osmate34水稻中MeHg含量,探究OsMATE34对水稻MeHg积累的影响。 结果 水稻OsMATE34基因位于8号染色体上,编码489个氨基酸,其编码蛋白的相对分子量为51.60 kD,理论等电点(isoelectric point, pI)为5.6,为疏水性蛋白。RT-qPCR结果显示,MeHg胁迫抑制OsMATE34在根部表达;叶鞘和叶中OsMATE34表达量随着MeHg浓度升高显著上调,其中80 nmol/L甲基汞处理时叶鞘中基因表达较对照上调20倍。随着MeHg浓度升高,水稻不同组织MeHg含量均呈上升趋势,根中MeHg含量远高于叶鞘和叶。水稻叶鞘和叶中OsMATE34基因的表达量与MeHg含量随着MeHg胁迫浓度升高呈显著的正相关关系。与野生型相比,OsMATE34敲除株系叶鞘、叶和木质部MeHg含量均显著下降,根中MeHg含量无显著差异。 结论 OsMATE34基因参与MeHg由根向地上部的转运,为后续研究水稻吸收转运MeHg的分子机制奠定基础。
杨跃琴, 邢英, 仲子荷, 田维军, 杨雪清, 王建旭. 甲基汞胁迫下水稻OsMATE34的表达及功能分析[J]. 生物技术通报, 2026, 42(1): 86-94.
YANG Yue-qin, XING Ying, ZHONG Zi-he, TIAN Wei-jun, YANG Xue-qing, WANG Jian-xu. Expression and Functional Analysis of OsMATE34 in Rice under Mercury Stress[J]. Biotechnology Bulletin, 2026, 42(1): 86-94.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| OsMATE34-qF | CATGAGACAAAGCGGCTGTG |
| OsMATE34-qR | TGAACATGGTGGTCACGGAG |
| OsActin-qF | GAGTATGATGAGTCGGGTCCAG |
| OsActin-qR | ACACCAACAATCCCAAACAGAG |
表1 RT-qPCR引物序列
Table 1 Primer sequence for RT-qPCR
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| OsMATE34-qF | CATGAGACAAAGCGGCTGTG |
| OsMATE34-qR | TGAACATGGTGGTCACGGAG |
| OsActin-qF | GAGTATGATGAGTCGGGTCCAG |
| OsActin-qR | ACACCAACAATCCCAAACAGAG |
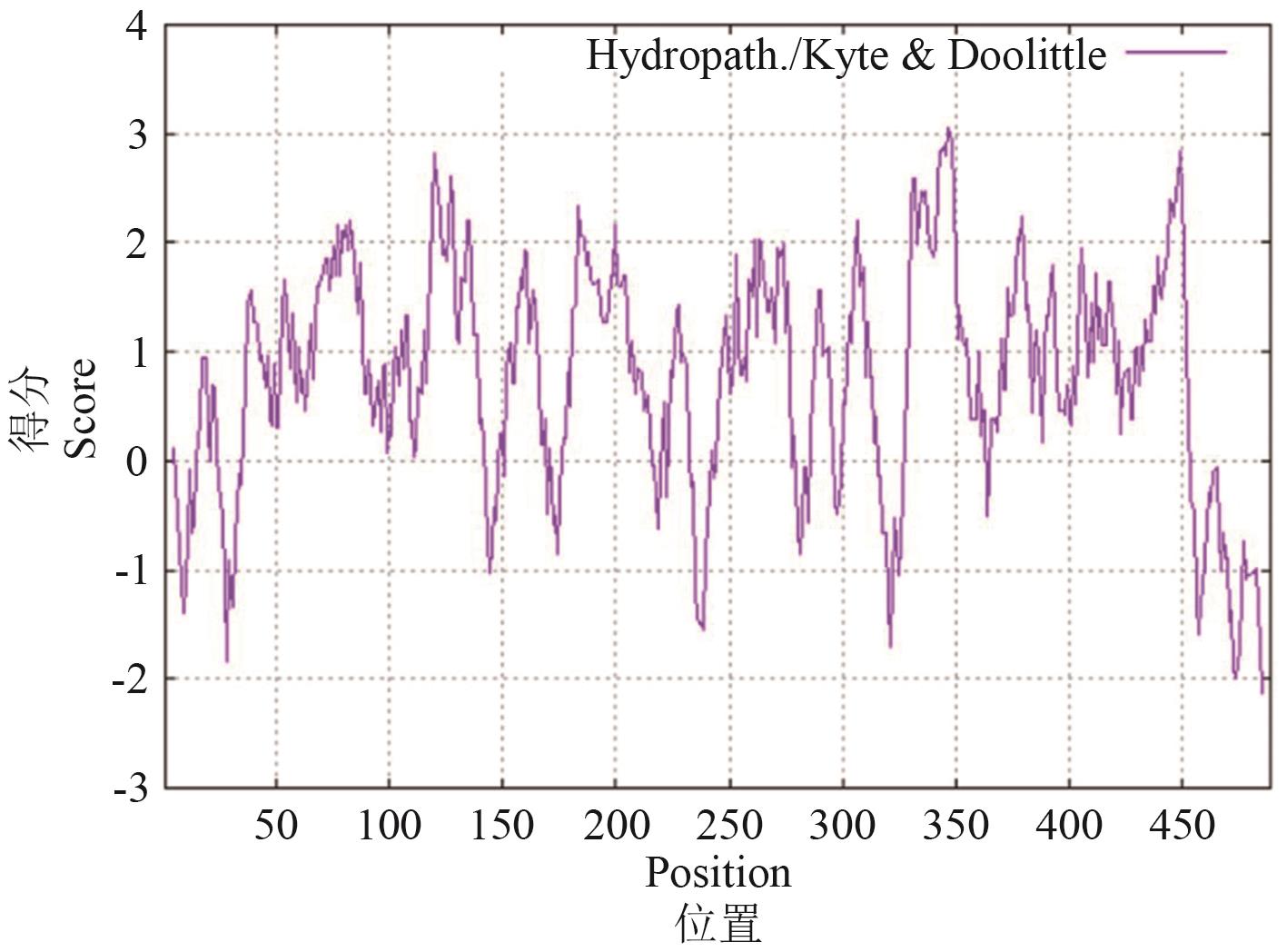
图1 OsMATE34蛋白亲/疏水性分析得分大于0的区域代表OsMATE34的疏水区,小于0的区域代表OsMATE34的亲水区
Fig. 1 Analysis of hydrophilicity/hydrophobicity of OsMATE34 proteinRegions with values above 0 are OsMATE34 hydrophobic in character. Regions with values less than 0 are OsMATE34 hydrophilic in character
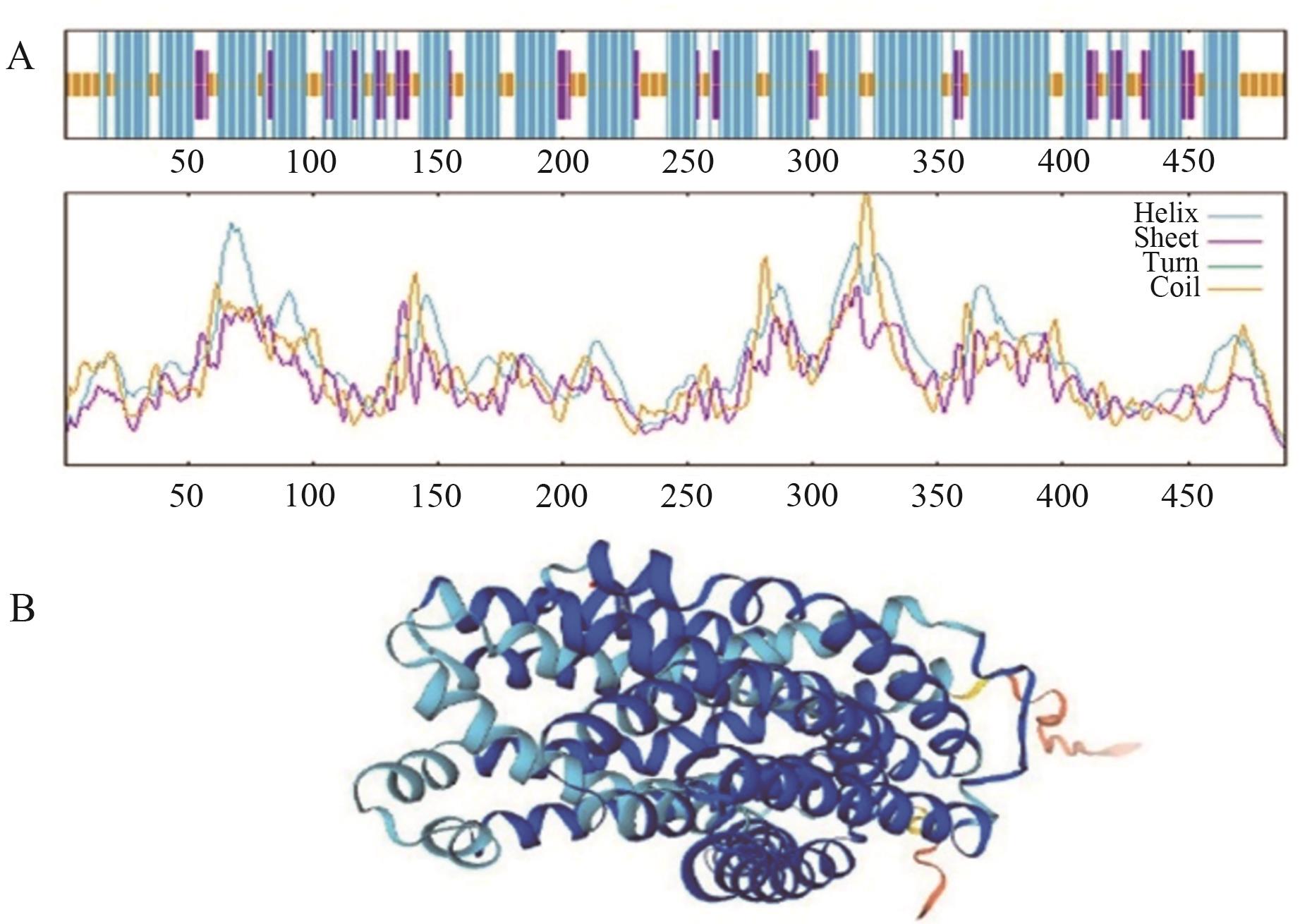
图2 OsMATE34蛋白质的二、三级结构预测A:OsMATE34的二级结构,蓝色表示α-螺旋,紫色表示延伸链,黄色表示无规则卷曲,横坐标表示每个氨基酸残基在OsMATE34上的位置;B:OsMATE34的三级结构,深蓝色表示α-螺旋,浅蓝色表示无规则卷曲,橙色表示延伸链
Fig. 2 Secondary and tertiary structure prediction of OsMATE34 proteinA: The secondary structure of OsMATE34, where blue indicates α-helix, purple indicates extended chain, yellow indicates irregular curl, and the abscissa indicates the position of each residue in the amino acid sequence of OsMATE34. B: The tertiary structure of OsMATE34, where dark blue indicates α-helix, light blue indicates irregular curl, and orange indicates extended chain

图4 OsMATE34蛋白的顺式作用元件分析不同颜色的方块代表生长素响应元件的一部分(蓝色)、脱落酸诱导顺式作用元件(深黄色)、厌氧诱导顺式作用元件(紫色)、生长素调控顺式作用元件(粉色)、茉莉酸甲酯诱导顺式作用元件(绿色)、参与玉米醇溶蛋白代谢调节的顺式作用调节元件(黄色)、与分生组织表达有关的顺式作用调控元件(浅绿色)、参与厌氧特异性诱导的增强子样元件(灰色)、赤霉素反应元件(浅黄色)、MYBHv1结合位点(浅粉色)
Fig. 4 Analysis of cis-acting elements of OsMATE34 proteinThe different colored squares indicate a portion of the growth hormone response element (blue), abscisic acid-induced cis-acting element (dark yellow), anaerobic-induced cis-acting element (purple), growth hormone-regulated cis-acting element (pink), methyl jasmonate-induced cis-acting element (green), cis-acting regulator involved in the regulation of zeinolysin metabolism (yellow), cis-acting related to the expression of meristematic tissue action regulatory element (light green), enhancer-like element involved in hypoxia-specific induction (gray), gibberellin-responsive element (light yellow), and MYBHv1 binding site (light pink)
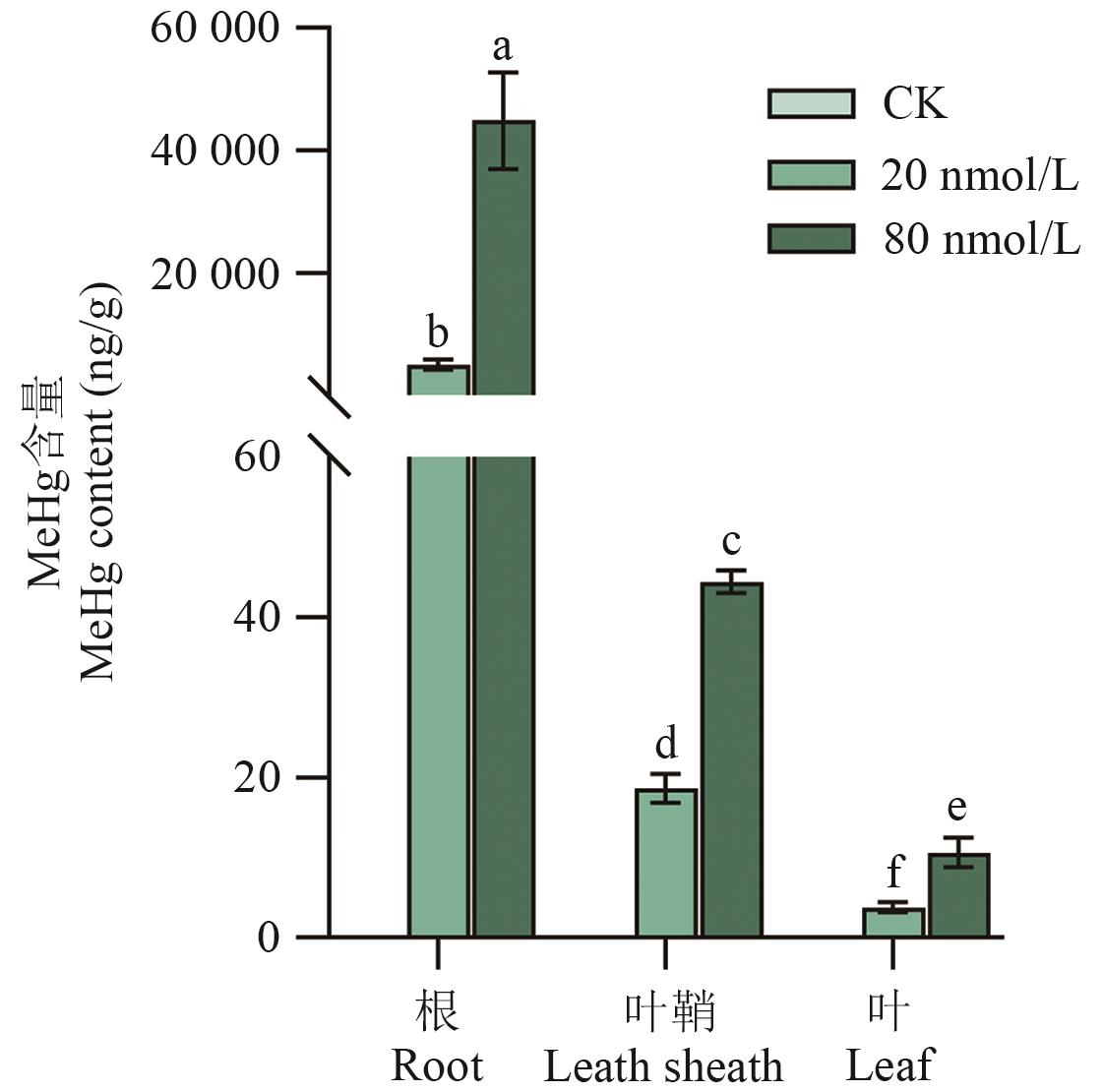
图6 水稻各组织中MeHg浓度变化不同小写字母表示差异显著(P<0.05)
Fig. 6 Variation in MeHg concentration in rice tissuesDifferent lowercase letters indicate significant differences between treatments (P<0.05)
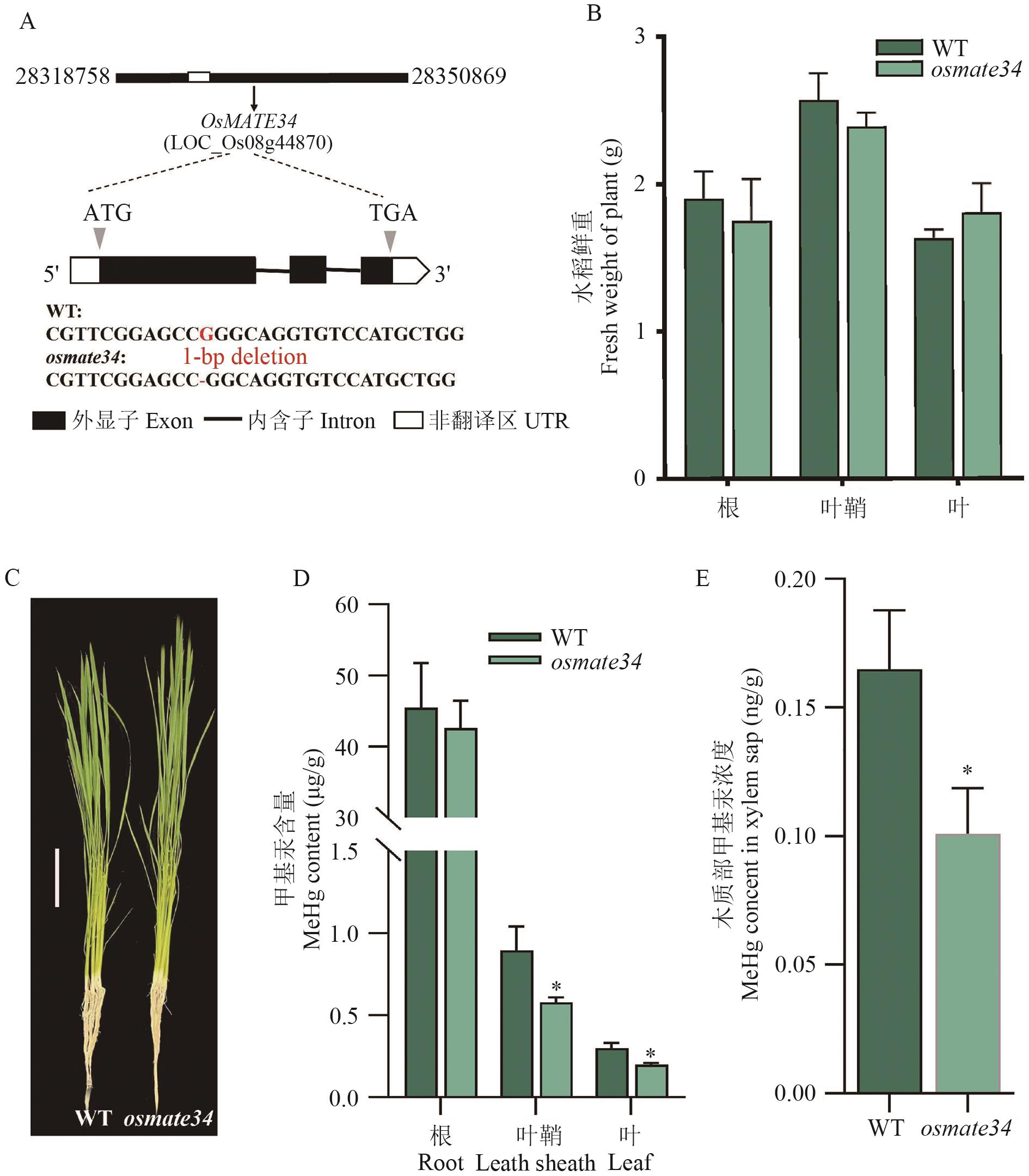
图8 osmate34和WT水培条件下的生长情况、MeHg含量A:OsMATE34突变靶标位点;B:osmate34和WT不同组织生物量;C:osmate34和WT的生长表型,比例尺=10 cm;D:osmate34和WT不同组织MeHg含量;E:osmate34和WT木质部中MeHg含量;* 表示相同条件下与野生型相比,0.05 水平上差异显著
Fig. 8 Growth conditions and MeHg contents of osmate34 and WT under hydroponic conditionsA: Mutated target site of OsMATE34 gene. B: Biomasses of osmate34 and WT in different tissues. C: Plant phenotypes of osmate34 and WT, scale =10 cm. D: MeHg contents of osmate34 and WT in different tissues. E: MeHg contents of osmate34 and WT in xylem sap. * indicates statistically significant difference in comparison to WT at P<0.05
| [1] | Axelrad DA, Bellinger DC, Ryan LM, et al. Dose-response relationship of prenatal mercury exposure and IQ: an integrative analysis of epidemiologic data [J]. Environ Health Perspect, 2007, 115(4): 609-615. |
| [2] | Wang B, Chen M, Ding L, et al. Fish, rice, and human hair mercury concentrations and health risks in typical Hg-contaminated areas and fish-rich areas, China [J]. Environ Int, 2021, 154: 106561. |
| [3] | Beckers F, Rinklebe J. Cycling of mercury in the environment: Sources, fate, and human health implications: a review [J]. Crit Rev Environ Sci Technol, 2017, 47(9): 693-794. |
| [4] | 陆本琦, 刘江, 吕文强, 等. 汞矿区稻田土壤汞形态分布特征及对甲基化的影响 [J]. 矿物岩石地球化学通报, 2021, 40(3): 690-698. |
| Lu BQ, Liu J, Lyu WQ, et al. Distribution characteristics of mercury occurrences in the paddy soil of Hg mining area and its effect on mercury methylation [J]. Bull Mineral Petrol Geochem, 2021, 40(3): 690-698. | |
| [5] | Zhang H, Feng XB, Larssen T, et al. Bioaccumulation of methylmercury versus inorganic mercury in rice (Oryza sativa L.) grain [J]. Environ Sci Technol, 2010, 44(12): 4499-4504. |
| [6] | Feng XB, Li P, Qiu GL, et al. Human exposure to methylmercury through rice intake in mercury mining areas, Guizhou province, China [J]. Environ Sci Technol, 2008, 42(1): 326-332. |
| [7] | Qiu GL, Feng XB, Li P, et al. Methylmercury accumulation in rice (Oryza sativa L.) grown at abandoned mercury mines in Guizhou, China [J]. J Agric Food Chem, 2008, 56(7): 2465-2468. |
| [8] | Li P, Feng XB, Chan HM, et al. Human body burden and dietary methylmercury intake: the relationship in a rice-consuming population [J]. Environ Sci Technol, 2015, 49(16): 9682-9689. |
| [9] | Meng B, Feng XB, Qiu GL, et al. The process of methylmercury accumulation in rice (Oryza sativa L.) [J]. Environ Sci Technol, 2011, 45(7): 2711-2717. |
| [10] | Hao YY, Zhu YJ, Yan RQ, et al. Important roles of thiols in methylmercury uptake and translocation by rice plants [J]. Environ Sci Technol, 2022, 56(10): 6765-6773. |
| [11] | Shoji T. ATP-binding cassette and multidrug and toxic compound extrusion transporters in plants [M]//International Review of Cell and Molecular Biology. Amsterdam: Elsevier, 2014: 303-346. |
| [12] | Huang Y, He GD, Tian WJ, et al. Genome-wide identification of MATE gene family in potato (Solanum tuberosum L.) and expression analysis in heavy metal stress [J]. Front Genet, 2021, 12: 650500. |
| [13] | Morita Y, Kodama K, Shiota S, et al. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticusand its homolog in Escherichia coli [J]. Antimicrob Agents Chemother, 1998, 42(7): 1778-1782. |
| [14] | Yang H, Guo D, Obianom ON, et al. Multidrug and toxin extrusion proteins mediate cellular transport of cadmium [J]. Toxicol Appl Pharmacol, 2017, 314: 55-62. |
| [15] | Lei GJ, Yokosho K, Yamaji N, et al. Two MATE transporters with different subcellular localization are involved in Al tolerance in buckwheat [J]. Plant Cell Physiol, 2017, 58(12): 2179-2189. |
| [16] | Vasconcellos RCC, Mendes FF, de Oliveira AC, et al. ZmMATE1 improves grain yield and yield stability in maize cultivated on acid soil [J]. Crop Sci, 2021, 61(5): 3497-3506. |
| [17] | Garcia-Oliveira AL, Martins-Lopes P, Tolrá R, et al. Molecular characterization of the citrate transporter gene TaMATE1 and expression analysis of upstream genes involved in organic acid transport under Al stress in bread wheat (Triticum aestivum) [J]. Physiol Plant, 2014, 152(3): 441-452. |
| [18] | Liu JP, Luo XY, Shaff J, et al. A promoter-swap strategy between the AtALMT and AtMATE genes increased Arabidopsis aluminum resistance and improved carbon-use efficiency for aluminum resistance [J]. Plant J, 2012, 71(2): 327-337. |
| [19] | Hoang MTT, Almeida D, Chay S, et al. AtDTX25, a member of the multidrug and toxic compound extrusion family, is a vacuolar ascorbate transporter that controls intracellular iron cycling in Arabidopsis [J]. New Phytol, 2021, 231(5): 1956-1967. |
| [20] | Mackon E, Ma YF, Jeazet Dongho Epse Mackon GC, et al. Computational and transcriptomic analysis unraveled OsMATE34 as a putative anthocyanin transporter in black rice (Oryza sativa L.) caryopsis [J]. Genes, 2021, 12(4): 583. |
| [21] | 王丽, 王万兴, 索海翠, 等. 马铃薯多酚氧化酶基因家族生物信息学及表达分析 [J]. 湖南农业大学学报: 自然科学版, 2019, 45(6): 601-610. |
| Wang L, Wang WX, Suo HC, et al. Bioinformatics and expression analysis of polyphenol oxidase gene family in potato [J]. J Hunan Agric Univ Nat Sci, 2019, 45(6): 601-610. | |
| [22] | Liang L, Horvat M, Cernichiari E, et al. Simple solvent extraction technique for elimination of matrix interferences in the determination of methylmercury in environmental and biological samples by ethylation-gas chromatography-cold vapor atomic fluorescence spectrometry [J]. Talanta, 1996, 43(11): 1883-1888. |
| [23] | Tang ZY, Fan FL, Wang XY, et al. Mercury in rice (Oryza sativa L.) and rice-paddy soils under long-term fertilizer and organic amendment [J]. Ecotoxicol Environ Saf, 2018, 150: 116-122. |
| [24] | 仇广乐, 冯新斌, 梁琏, 等. 溶剂萃取-水相乙基化衍生GC-CVAFS联用测定苔藓样品中的甲基汞 [J]. 分析测试学报, 2005, 24(1): 29-32. |
| Qiu GL, Feng XB, Liang L, et al. Determination of methylmercury in moss by ethylation-gas chromatography-cold vapor atomic fluorescence spectrometry with solvent extraction [J]. J Instrum Anal, 2005, 24(1): 29-32. | |
| [25] | Ikai A. Thermostability and aliphatic index of globular proteins [J]. The Journal of Biochemistry, 1980, 88(6): 1895-1898. |
| [26] | Yazaki K, Sugiyama A, Morita M, et al. Secondary transport as an efficient membrane transport mechanism for plant secondary metabolites [J]. Phytochem Rev, 2008, 7(3): 513-524. |
| [27] | Colmenero-Flores JM, Franco-Navarro JD, Cubero-Font P, et al. Chloride as a beneficial macronutrient in higher plants: new roles and regulation [J]. Int J Mol Sci, 2019, 20(19): 4686. |
| [28] | Inoue SI, Hayashi M, Huang S, et al. A tonoplast-localized magnesium transporter is crucial for stomatal opening in Arabidopsis under high Mg2+ conditions [J]. New Phytol, 2022, 236(3): 864-877. |
| [29] | 方波, 肖腾伟, 苏娜娜, 等. 水稻镉吸收及其在各器官间转运积累的研究进展 [J]. 中国水稻科学, 2021, 35(3): 225-237. |
| Fang B, Xiao TW, Su NN, et al. Research progress on cadmium uptake and its transport and accumulation among organs in rice [J]. Chin J Rice Sci, 2021, 35(3): 225-237. | |
| [30] | 许肖博, 安鹏虎, 郭天骄, 等. 水稻镉胁迫响应机制及防控措施研究进展 [J]. 中国水稻科学, 2021, 35(5): 415-426. |
| Xu XB, An PH, Guo TJ, et al. Research progresses on response mechanisms and control measures of cadmium stress in rice [J]. Chin J Rice Sci, 2021, 35(5): 415-426. | |
| [31] | Li NN, Meng HJ, Xing HT, et al. Genome-wide analysis of MATE transporters and molecular characterization of aluminum resistance in Populus [J]. J Exp Bot, 2017, 68(20): 5669-5683. |
| [32] | Wang JJ, Hou QQ, Li PH, et al. Diverse functions of multidrug and toxin extrusion (MATE) transporters in citric acid efflux and metal homeostasis in Medicago truncatula [J]. Plant J, 2017, 90(1): 79-95. |
| [33] | Li DD, Xu XM, Hu XQ, et al. Genome-wide analysis and heavy metal-induced expression profiling of the HMA gene family in Populus trichocarpa [J]. Front Plant Sci, 2015, 6: 1149. |
| [34] | Strickman RJ, Mitchell CPJ. Accumulation and translocation of methylmercury and inorganic mercury in Oryza sativa: an enriched isotope tracer study [J]. Sci Total Environ, 2017, 574: 1415-1423. |
| [35] | Beauvais-Flück R, Slaveykova VI, Cosio C. Effects of two-hour exposure to environmental and high concentrations of methylmercury on the transcriptome of the macrophyte Elodea nuttallii [J]. Aquat Toxicol, 2018, 194: 103-111. |
| [36] | Kengo Yokosho NY. OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice [J]. Plant Physiol, 2009, 149(1): 297-305. |
| [37] | Das N, Bhattacharya S, Bhattacharyya S, et al. Expression of rice MATE family transporter OsMATE2 modulates arsenic accumulation in tobacco and rice [J]. Plant Mol Biol, 2018, 98(1): 101-120. |
| [38] | 张永福, 莫丽玲, 徐金会, 等. 葡萄耐铝毒基因MATE的克隆与表达分析 [J]. 江西农业大学学报, 2020, 42(5): 906-914. |
| Zhang YF, Mo LL, Xu JH, et al. Cloning and expression analysis of the aluminum-toxicity-tolerance gene MATE in grape [J]. Acta Agric Univ Jiangxiensis, 2020, 42(5): 906-914. | |
| [39] | Yokosho K, Yamaji N, Ma JF. OsFRDL1 expressed in nodes is required for distribution of iron to grains in rice [J]. J Exp Bot, 2016, 67(18): 5485-5494. |
| [1] | 任云儿, 伍国强, 成斌, 魏明. 甜菜BvATGs基因家族全基因组鉴定及盐胁迫下表达模式分析[J]. 生物技术通报, 2026, 42(1): 184-197. |
| [2] | 费思恬, 侯鹰翔, 李兰, 张超. 水稻赤霉素信号负调控因子SLR1的生物学功能及其调控网络[J]. 生物技术通报, 2026, 42(1): 13-30. |
| [3] | 陈静欢, 房国楠, 朱文豪, 叶广继, 苏旺, 贺苗苗, 杨生龙, 周云. 马铃薯种质资源淀粉表征及相关基因表达分析[J]. 生物技术通报, 2026, 42(1): 170-183. |
| [4] | 王芳, 邵会茹, 吕林龙, 赵点, 胡振, 吕建珍, 姜亮. 植物和细菌TurboID邻近蛋白标记方法的建立[J]. 生物技术通报, 2025, 41(9): 44-53. |
| [5] | 李珊, 马登辉, 马红义, 姚文孔, 尹晓. 葡萄SKP1基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(9): 147-158. |
| [6] | 巩慧玲, 邢玉洁, 马俊贤, 蔡霞, 冯再平. 马铃薯LAC基因家族的鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2025, 41(9): 82-93. |
| [7] | 化文平, 刘菲, 浩佳欣, 陈尘. 丹参ADH基因家族的鉴定与表达模式分析[J]. 生物技术通报, 2025, 41(8): 211-219. |
| [8] | 邓美壁, 严浪, 詹志田, 朱敏, 和玉兵. RUBY辅助的水稻高效CRISPR基因编辑[J]. 生物技术通报, 2025, 41(8): 65-73. |
| [9] | 白雨果, 李婉迪, 梁建萍, 石志勇, 卢庚龙, 刘红军, 牛景萍. 哈茨木霉T9131对黄芪幼苗的促生机理[J]. 生物技术通报, 2025, 41(8): 175-185. |
| [10] | 任睿斌, 司二静, 万广有, 汪军成, 姚立蓉, 张宏, 马小乐, 李葆春, 王化俊, 孟亚雄. 大麦条纹病菌GH17基因家族的鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 146-154. |
| [11] | 曾丹, 黄园, 王健, 张艳, 刘庆霞, 谷荣辉, 孙庆文, 陈宏宇. 铁皮石斛bZIP转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 197-210. |
| [12] | 侯鹰翔, 费思恬, 黎妮, 李兰, 宋松泉, 王伟平, 张超. 水稻miRNAs响应生物胁迫研究进展[J]. 生物技术通报, 2025, 41(7): 69-80. |
| [13] | 李凯月, 邓晓霞, 殷缘, 杜亚彤, 徐元静, 王竞红, 于耸, 蔺吉祥. 蓖麻LEA基因家族的鉴定和铝胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 128-138. |
| [14] | 牛景萍, 赵婧, 郭茜, 王书宏, 赵晋忠, 杜维俊, 殷丛丛, 岳爱琴. 基于WGCNA鉴定大豆抗大豆花叶病毒NAC转录因子及其诱导表达分析[J]. 生物技术通报, 2025, 41(7): 95-105. |
| [15] | 韩燚, 侯昌林, 唐露, 孙璐, 谢晓东, 梁晨, 陈小强. 大麦HvERECTA基因的克隆及功能分析[J]. 生物技术通报, 2025, 41(7): 106-116. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||