








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (11): 248-256.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1348
Previous Articles Next Articles
LIU Wen-fu( ), HUANG Dan-qiong, HU Qun-ju, WANG Chao-gang(
), HUANG Dan-qiong, HU Qun-ju, WANG Chao-gang( )
)
Received:2020-11-03
Online:2021-11-26
Published:2021-12-03
Contact:
WANG Chao-gang
E-mail:2170257312@email.szu.edu.cn;charles@szu.edu.cn
LIU Wen-fu, HUANG Dan-qiong, HU Qun-ju, WANG Chao-gang. Acquisition and Expression Analysis of the Transcription Factor Whirly1 from Haematococcus pluvialis[J]. Biotechnology Bulletin, 2021, 37(11): 248-256.

Fig.6 Homology analysis of HpWhirly1 and other WHYs from different species Haematococcus lacustris(GFH09517.1),Chlamydomonas eustigma(GAX74163.1),Dunaliella salina(KAF5835724.1),and Micractinium conductrix(PSC72124.1). Three Whirly transcription factor sequences in the model organism Arabidopsis:AtWHY1(AT1G14410),AtWHY2(AT1G71260),and AtWHY3(AT2G02740)
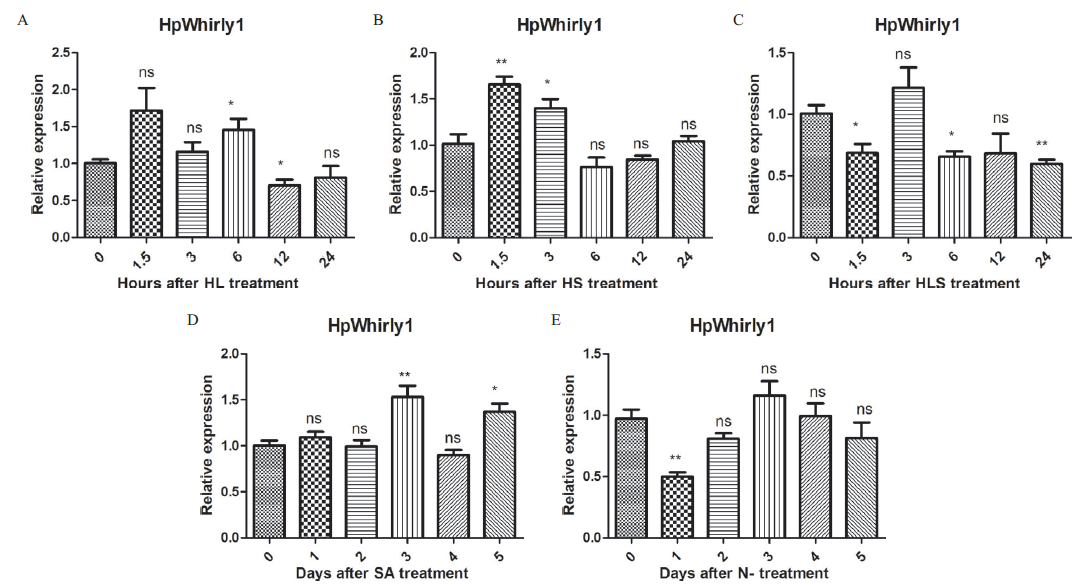
Fig.8 Expression analysis of HpWhirly1 under different stress conditions A: Strong light treatment. B: Acetic acid treatment. C: Strong light sodium acetate combined treatment. D: Salicylic acid treatment. E: Nitrogen deficiency treatment. * indicates significant difference between treatment group and control group(P<0.05). ** indicates extremely significant difference between treatment group and control group(P<0.01)
| [1] |
Lee YK, Ding SY. Cell cycle and accumulation of astaxanthin in Haematococcus lacustris(chlorophyta)1[J]. J Phycol, 1994, 30(3): 445-449.
doi: 10.1111/j.0022-3646.1994.00445.x URL |
| [2] |
Chamovitz D, Sandmann G, Hirschberg J. Molecular and biochemical characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate-limiting step in carotenoid biosynjournal[J]. J Biol Chem, 1993, 268(23): 17348-17353.
pmid: 8349618 |
| [3] |
Steinbrenner J, Linden H. Regulation of two carotenoid biosynjournal genes coding for phytoene synthase and carotenoid hydroxylase during stress-induced astaxanthin formation in the green alga Haematococcus pluvialis[J]. Plant Physiol, 2001, 125(2): 810-817.
pmid: 11161038 |
| [4] | 王坤鹏. 雨生红球藻的转录组测序及乙酰转移酶基因的克隆和分析[D]. 深圳:深圳大学, 2017. |
| Wang KP. Transcriptome sequencing of haematoccus Pluvialis and cloning and analysis of the acetyltransferase gene[D]. Shenzhen:Shenzhen University, 2017. | |
| [5] | Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses[J]. Biochim Biophys Acta, 2012, 1819(2): 86-96. |
| [6] |
Dickey TH, Altschuler SE, Wuttke DS. Single-stranded DNA-binding proteins:multiple domains for multiple functions[J]. Structure, 2013, 21(7): 1074-1084.
doi: 10.1016/j.str.2013.05.013 URL |
| [7] | 姚沁涛, 张文蔚, 刘莉, 等. Whirly转录因子对非寄主菌诱导水稻HR反应的负调控作用[J]. 中国农业科技导报, 2008, 10(5): 53-58. |
| Yao QT, Zhang WW, Liu L, et al. Negative regulation of rice whirly transcription factor for the hypersensitive response induced by non-host pathogen bacterium[J]. J Agric Sci Technol, 2008, 10(5): 53-58. | |
| [8] | 姚侠妹. 侧柏古树端粒相关基因克隆及其抗逆功能研究[D]. 北京:中国林业科学研究院, 2017. |
| Yao XM. Identification of telomere-related genes from old Platycladus orientalis and their involvement in abiotic stress tolerance[D]. Beijing:Chinese Academy of Forestry, 2017. | |
| [9] |
Zhang YF, Hou MM, Tan BC. The requirement of WHIRLY1 for embryogenesis is dependent on genetic background in maize[J]. PLoS One, 2013, 8(6): e67369.
doi: 10.1371/journal.pone.0067369 URL |
| [10] |
Isemer R, Krause K, Grabe N, et al. Plastid located WHIRLY1 enhances the responsiveness of Arabidopsis seedlings toward abscisic acid[J]. Front Plant Sci, 2012, 3: 283.
doi: 10.3389/fpls.2012.00283 pmid: 23269926 |
| [11] | 蔡倩, 任育军, 黄晨星, 等. 植物单链DNA结合蛋白WHIRLY2研究进展[J]. 福建农林大学学报:自然科学版, 2018, 47(3): 267-273. |
| Cai Q, Ren YJ, Huang CX, et al. Research progress of single-stranded DNA binding protein WHIRLY2 in plants[J]. J Fujian Agric For Univ:Nat Sci Ed, 2018, 47(3): 267-273. | |
| [12] |
Yoo HH, Kwon C, Lee MM, et al. Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis[J]. Plant J, 2007, 49(3): 442-451.
doi: 10.1111/tpj.2007.49.issue-3 URL |
| [13] |
Xiong JY, Lai CX, Qu Z, et al. Recruitment of AtWHY1 and AtWHY3 by a distal element upstream of the kinesin gene AtKP1 to mediate transcriptional repression[J]. Plant Mol Biol, 2009, 71(4/5): 437-449.
doi: 10.1007/s11103-009-9533-7 URL |
| [14] |
Miao Y, Jiang J, Ren Y, et al. The single-stranded DNA-binding protein WHIRLY1 represses WRKY53 expression and delays leaf senescence in a developmental stage-dependent manner in Arabidopsis[J]. Plant Physiol, 2013, 163(2): 746-756.
doi: 10.1104/pp.113.223412 URL |
| [15] |
Krause K, Kilbienski I, Mulisch M, et al. DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles[J]. FEBS Lett, 2005, 579(17): 3707-3712.
doi: 10.1016/j.febslet.2005.05.059 URL |
| [16] |
Desveaux D, Allard J, Brisson N, et al. A new family of plant transcription factors displays a novel ssDNA-binding surface[J]. Nat Struct Biol, 2002, 9(7): 512-517.
pmid: 12080340 |
| [17] |
Desveaux D, Subramaniam R, Després C, et al. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis[J]. Dev Cell, 2004, 6(2): 229-240.
pmid: 14960277 |
| [18] |
Desveaux D, Maréchal A, Brisson N. Whirly transcription factors:defense gene regulation and beyond[J]. Trends Plant Sci, 2005, 10(2): 95-102.
pmid: 15708347 |
| [19] |
Desveaux D, Després C, Joyeux A, et al. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato[J]. Plant Cell, 2000, 12(8): 1477-1489.
pmid: 10948264 |
| [20] | 王洁如. 番茄LeWhy1低温胁迫下的功能分析及转录组分析[D]. 泰安:山东农业大学, 2015. |
| Wang JR. Functional analysis and RNA-sequencing of LeWhy1 in tomato under chilling stress[D]. Tai’an:Shandong Agricultural University, 2015. | |
| [21] | 林文芳, 任育军, 缪颖. 植物Whirly蛋白调控叶片衰老的研究进展[J]. 植物生理学报, 2014, 50(9): 1274-1284. |
| Lin WF, Ren YJ, Miao Y. Research progress of whirly proteins in regulation of leaf senescence[J]. Plant Physiol J, 2014, 50(9): 1274-1284. | |
| [22] |
Cappadocia L, Parent JS, Zampini E, et al. A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage[J]. Nucleic Acids Res, 2012, 40(1): 258-269.
doi: 10.1093/nar/gkr740 pmid: 21911368 |
| [23] |
Grabowski E, Miao Y, Mulisch M, et al. Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell[J]. Plant Physiol, 2008, 147(4): 1800-1804.
doi: 10.1104/pp.108.122796 pmid: 18678751 |
| [24] |
Isemer R, Mulisch M, Schäfer A, et al. Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus[J]. FEBS Lett, 2012, 586(1): 85-88.
doi: 10.1016/j.febslet.2011.11.029 pmid: 22154598 |
| [25] |
Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology[J]. Nucleic Acids Res, 2002, 30(6): 1292-1305.
pmid: 11884626 |
| [26] |
Cappadocia L, Sygusch J, Brisson N. Purification, crystallization and preliminary X-ray diffraction analysis of the Whirly domain of StWhy2 in complex with single-stranded DNA[J]. Acta Crystallogr Sect F Struct Biol Cryst Commun, 2008, 64(pt 11): 1056-1059.
doi: 10.1107/S1744309108032399 pmid: 18997341 |
| [27] |
Cappadocia L, Parent JS, Sygusch J, et al. A family portrait:structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum[J]. Acta Crystallogr Sect F Struct Biol Cryst Commun, 2013, 69(pt 11): 1207-1211.
doi: 10.1107/S1744309113028698 pmid: 24192350 |
| [28] |
Cappadocia L, Maréchal A, Parent JS, et al. Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair[J]. Plant Cell, 2010, 22(6): 1849-1867.
doi: 10.1105/tpc.109.071399 URL |
| [29] |
Maréchal A, Parent JS, Sabar M, et al. Overexpression of mtDNA-associated AtWhy2 compromises mitochondrial function[J]. BMC Plant Biol, 2008, 8: 42.
doi: 10.1186/1471-2229-8-42 URL |
| [30] | 董庆霖, 赵学明, 邢向英. 乙酸钠诱导雨生红球藻合成虾青素的机理[J]. 微生物学通报, 2007, 34(2): 256-260. |
| Dong QL, Zhao XM, Xing XY. Inducing mechanism of acetate on astaxanthin synjournal in Haematococcus pluvialis[J]. Microbiology, 2007, 34(2): 256-260. | |
| [31] |
Gao ZQ, Miao XX, Zhang XW, et al. Comparative fatty acid transcriptomic test and iTRAQ-based proteomic analysis in Haematococcus pluvialis upon salicylic acid(SA)and jasmonic acid(JA)inductions[J]. Algal Res, 2016, 17: 277-284.
doi: 10.1016/j.algal.2016.05.012 URL |
| [32] |
Griffiths MJ, Harrison STL. Lipid productivity as a key characteristic for choosing algal species for biodiesel production[J]. J Appl Phycol, 2009, 21(5): 493-507.
doi: 10.1007/s10811-008-9392-7 URL |
| [33] |
Solovchenko AE. Recent breakthroughs in the biology of astaxanthin accumulation by microalgal cell[J]. Photosynth Res, 2015, 125(3): 437-449.
doi: 10.1007/s11120-015-0156-3 URL |
| [34] |
Mascia F, Girolomoni L, Alcocer MJP, et al. Functional analysis of photosynthetic pigment binding complexes in the green alga Haematococcus pluvialis reveals distribution of astaxanthin in Photosystems[J]. Sci Rep, 2017, 7(1): 16319.
doi: 10.1038/s41598-017-16641-6 URL |
| [1] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [2] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [3] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [4] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [5] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [6] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [7] | GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L. [J]. Biotechnology Bulletin, 2023, 39(6): 217-232. |
| [8] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [9] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [10] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| [11] | GE Yan-rui, ZHAO Ran, XU Jing, LI Ruo-fan, HU Yun-tao, LI Rui-li. Advances in the Development and Regulation of Vascular Cambium [J]. Biotechnology Bulletin, 2023, 39(3): 13-25. |
| [12] | LIU Cheng-xia, SUN Zong-yan, LUO Yun-bo, ZHU Hong-liang, QU Gui-qin. Multifaceted Roles of bHLH Phosphorylation in Regulation of Plant Physiological Functions [J]. Biotechnology Bulletin, 2023, 39(3): 26-34. |
| [13] | ZHAO Meng-liang, GUO Yi-ting, REN Yan-jing. Identification and Analysis of WRKY Transcription Factor Family Genes in Helianthus tuberosus [J]. Biotechnology Bulletin, 2023, 39(2): 116-125. |
| [14] | HAN Fang-ying, HU Xin, WANG Nan-nan, XIE Yu-hong, WANG Xiao-yan, ZHU Qiang. Research Progress in Response of DREBs to Abiotic Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(11): 86-98. |
| [15] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||