








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (12): 1-12.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0184
ZHAN Dong-mei1( ), ZHU Chen1,2, ZHOU Cheng-zhe1,2, HUANG Xue-ting1, LAI Zhong-xiong1,2, GUO Yu-qiong1,2(
), ZHU Chen1,2, ZHOU Cheng-zhe1,2, HUANG Xue-ting1, LAI Zhong-xiong1,2, GUO Yu-qiong1,2( )
)
Received:2021-02-14
Online:2021-12-26
Published:2022-01-19
Contact:
GUO Yu-qiong
E-mail:dongmeizhan@foxmail.com;guoyq828@163.com
ZHAN Dong-mei, ZHU Chen, ZHOU Cheng-zhe, HUANG Xue-ting, LAI Zhong-xiong, GUO Yu-qiong. Genome-wide Identification of Gro/Tup1 Gene Family in Camellia sinensis and Their Expression Analysis Under Exogenous Phytohormones and Abiotic Stress Treatments[J]. Biotechnology Bulletin, 2021, 37(12): 1-12.
| 基因名称 Gene name | 上游引物 Upstream primer | 下游引物 Downstream primer |
|---|---|---|
| CsLUG1 | GCGAGTCATCTGGGTAGTAAGT | GTTGTTGGTGCTCTCTTGCT |
| CsLUG2 | GCTGAGAGTGCTTTGGATGG | GCTGAAGGATGTTGAGTGCT |
| CsLUG3 | TTGGAATGCGAGTGGTCAGT | TGGTGCTGTGGTGTCAACAT |
| CsLUG4 | CGTGGCAAGCCCTTAAAG | TGGTGTCAGCATCTGGAGT |
| CsTPL1 | CATGTGTTGAAGAGACTGAGG | TGGGCAAATCATCAGAAGAG |
| CsTPL2 | GCTACCGCTGTTGAGATTCTTG | ATTTGCCAGGGTGAGGAGTAG |
| CsTPL3 | CCTTCCAACCTACACCAGC | CACCAAGACCAATAGCTCCAC |
| CsTPL4 | TCGGCTTTGTCAGTTCTACG | CCGGAAGTGTTATAGGAGGTG |
| CsTPL5 | GCGACTTGAGCATCTCCTTG | GCTGCCACTTCAAAGCCT |
| CsTPL6 | TGGCTCGCTCTCCATGTATG | TGCAAGAGGAACGTGATGGT |
| CsTPL7 | AGATGTTGGTTTGGTTCAGGG | GCTTTGGTTAATGAGGTTCCG |
| CsTPL8 | TTCAAGGTTGCGGACTCTCAT | GATTGTTTGCTGGTGATGGAG |
| CsTPL9 | GTCGCCTGTAACTAACCCACT | TGAGGAACCGGAGATGGATTAG |
| Csβ-actin | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT |
| CsSAND | CCAATTGCCCCCTTAATGAC | GCAATCATTTCCTTCGTGGAG |
Table 1 Primer sequences used in RT-qPCR
| 基因名称 Gene name | 上游引物 Upstream primer | 下游引物 Downstream primer |
|---|---|---|
| CsLUG1 | GCGAGTCATCTGGGTAGTAAGT | GTTGTTGGTGCTCTCTTGCT |
| CsLUG2 | GCTGAGAGTGCTTTGGATGG | GCTGAAGGATGTTGAGTGCT |
| CsLUG3 | TTGGAATGCGAGTGGTCAGT | TGGTGCTGTGGTGTCAACAT |
| CsLUG4 | CGTGGCAAGCCCTTAAAG | TGGTGTCAGCATCTGGAGT |
| CsTPL1 | CATGTGTTGAAGAGACTGAGG | TGGGCAAATCATCAGAAGAG |
| CsTPL2 | GCTACCGCTGTTGAGATTCTTG | ATTTGCCAGGGTGAGGAGTAG |
| CsTPL3 | CCTTCCAACCTACACCAGC | CACCAAGACCAATAGCTCCAC |
| CsTPL4 | TCGGCTTTGTCAGTTCTACG | CCGGAAGTGTTATAGGAGGTG |
| CsTPL5 | GCGACTTGAGCATCTCCTTG | GCTGCCACTTCAAAGCCT |
| CsTPL6 | TGGCTCGCTCTCCATGTATG | TGCAAGAGGAACGTGATGGT |
| CsTPL7 | AGATGTTGGTTTGGTTCAGGG | GCTTTGGTTAATGAGGTTCCG |
| CsTPL8 | TTCAAGGTTGCGGACTCTCAT | GATTGTTTGCTGGTGATGGAG |
| CsTPL9 | GTCGCCTGTAACTAACCCACT | TGAGGAACCGGAGATGGATTAG |
| Csβ-actin | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT |
| CsSAND | CCAATTGCCCCCTTAATGAC | GCAATCATTTCCTTCGTGGAG |
| 基因名称 Gene name | 基因ID Gene ID | 大小 Size/aa | 分子量 Molecular weight/kD | 等电点 pI | 亚细胞定位 Subcellular localization | 不稳定系数 Instability index | 脂溶指 Aliphatic index | 亲水指数 GRAVY | 外显子数 Number of exons |
|---|---|---|---|---|---|---|---|---|---|
| CsLUG1 | TEA022439.1 | 864 | 94.51266 | 7.02 | 质体Plastid | 46.11 | 77.78 | 0.342 | 17 |
| CsLUG2 | TEA004970.1 | 660 | 73.86218 | 8.04 | 细胞核Nucleus | 45.09 | 67.7 | 0.566 | 16 |
| CsLUG3 | TEA020168.1 | 907 | 100.83071 | 6.48 | 细胞核Nucleus | 53.22 | 67.42 | 0.648 | 19 |
| CsLUG4 | TEA025268.1 | 757 | 81.54457 | 6.08 | 细胞核Nucleus | 40.86 | 40.86 | 0.352 | 17 |
| CsTPL1 | TEA031994.1 | 540 | 60.18248 | 8.07 | 细胞核Nucleus | 37.53 | 79.65 | 0.368 | 14 |
| CsTPL2 | TEA019471.1 | 1196 | 131.4561 | 6.33 | 细胞核Nucleus | 40.35 | 83.09 | 0.27 | 27 |
| CsTPL3 | TEA016267.1 | 1319 | 145.85055 | 6.87 | 细胞核Nucleus | 39.33 | 77.87 | 0.317 | 30 |
| CsTPL4 | TEA030187.1 | 569 | 64.03845 | 6.27 | 叶绿体Chloroplast | 39.24 | 91.92 | 0.193 | 6 |
| CsTPL5 | TEA024264.1 | 723 | 81.24096 | 8.17 | 细胞核Nucleus | 43.53 | 85.01 | 0.244 | 20 |
| CsTPL6 | TEA022067.1 | 1069 | 118.97318 | 6.14 | 内质网Reticulum | 39.47 | 86.43 | 0.222 | 24 |
| CsTPL7 | TEA022077.1 | 759 | 86.17475 | 6.72 | 细胞核Nucleus | 43.25 | 85.03 | 0.181 | 18 |
| CsTPL8 | TEA020620.1 | 1195 | 131.50275 | 6.43 | 叶绿体Chloroplast | 40.03 | 80.14 | 0.312 | 26 |
| CsTPL9 | TEA023424.1 | 1188 | 131.09747 | 6.3 | 细胞核Nucleus | 38.29 | 79.55 | 0.317 | 26 |
Table 2 Feature of CsGro/Tup1 protein in C. sinensis
| 基因名称 Gene name | 基因ID Gene ID | 大小 Size/aa | 分子量 Molecular weight/kD | 等电点 pI | 亚细胞定位 Subcellular localization | 不稳定系数 Instability index | 脂溶指 Aliphatic index | 亲水指数 GRAVY | 外显子数 Number of exons |
|---|---|---|---|---|---|---|---|---|---|
| CsLUG1 | TEA022439.1 | 864 | 94.51266 | 7.02 | 质体Plastid | 46.11 | 77.78 | 0.342 | 17 |
| CsLUG2 | TEA004970.1 | 660 | 73.86218 | 8.04 | 细胞核Nucleus | 45.09 | 67.7 | 0.566 | 16 |
| CsLUG3 | TEA020168.1 | 907 | 100.83071 | 6.48 | 细胞核Nucleus | 53.22 | 67.42 | 0.648 | 19 |
| CsLUG4 | TEA025268.1 | 757 | 81.54457 | 6.08 | 细胞核Nucleus | 40.86 | 40.86 | 0.352 | 17 |
| CsTPL1 | TEA031994.1 | 540 | 60.18248 | 8.07 | 细胞核Nucleus | 37.53 | 79.65 | 0.368 | 14 |
| CsTPL2 | TEA019471.1 | 1196 | 131.4561 | 6.33 | 细胞核Nucleus | 40.35 | 83.09 | 0.27 | 27 |
| CsTPL3 | TEA016267.1 | 1319 | 145.85055 | 6.87 | 细胞核Nucleus | 39.33 | 77.87 | 0.317 | 30 |
| CsTPL4 | TEA030187.1 | 569 | 64.03845 | 6.27 | 叶绿体Chloroplast | 39.24 | 91.92 | 0.193 | 6 |
| CsTPL5 | TEA024264.1 | 723 | 81.24096 | 8.17 | 细胞核Nucleus | 43.53 | 85.01 | 0.244 | 20 |
| CsTPL6 | TEA022067.1 | 1069 | 118.97318 | 6.14 | 内质网Reticulum | 39.47 | 86.43 | 0.222 | 24 |
| CsTPL7 | TEA022077.1 | 759 | 86.17475 | 6.72 | 细胞核Nucleus | 43.25 | 85.03 | 0.181 | 18 |
| CsTPL8 | TEA020620.1 | 1195 | 131.50275 | 6.43 | 叶绿体Chloroplast | 40.03 | 80.14 | 0.312 | 26 |
| CsTPL9 | TEA023424.1 | 1188 | 131.09747 | 6.3 | 细胞核Nucleus | 38.29 | 79.55 | 0.317 | 26 |

Fig. 3 Gro/Tup1 in C. sinensis and A. thaliana A:Sequence evolution relationship of Gro/Tup1 protein. B:Conservative motif distribution of Gro/Tup1. C:Gro/Tup1 gene intron-exon structure distribution
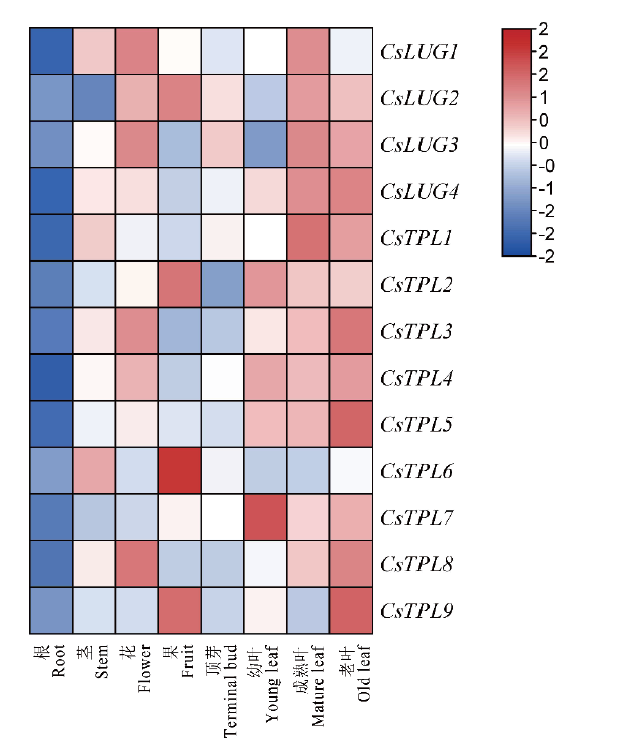
Fig.6 Expression profiles of CsGro/Tup1 family genes in the tissues of ‘Tieguanyin’ cultivar The color scale is the log2 converted value. Red indicates high expression,and blue indicates low expression
| [1] |
Courey AJ, Jia S. Transcriptional repression:the long and the short of it[J]. Genes Dev, 2001, 15(21):2786-2796.
doi: 10.1101/gad.939601 URL |
| [2] |
Krogan NT, Long JA. Why so repressed? Turning off transcription during plant growth and development[J]. Curr Opin Plant Biol, 2009, 12(5):628-636.
doi: 10.1016/j.pbi.2009.07.011 pmid: 19700365 |
| [3] |
Mannervik M, Nibu Y, Zhang H, et al. Transcriptional coregulators in development[J]. Science, 1999, 284(5414):606-609.
pmid: 10213677 |
| [4] |
Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression[J]. Gene, 2000, 249(1/2):1-16.
doi: 10.1016/S0378-1119(00)00161-X URL |
| [5] |
Conner J, Liu Z. LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development[J]. PNAS, 2000, 97(23):12902-12907.
pmid: 11058164 |
| [6] |
Long JA, Ohno C, Smith ZR, et al. TOPLESS regulates apical embryonic fate in Arabidopsis[J]. Science, 2006, 312(5779):1520-1523.
doi: 10.1126/science.1123841 URL |
| [7] |
Kieffer M, Stern Y, Cook H, et al. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance[J]. Plant Cell, 2006, 18(3):560-573.
pmid: 16461579 |
| [8] |
Liu ZC, Karmarkar V. Groucho/Tup1 family co-repressors in plant development[J]. Trends Plant Sci, 2008, 13(3):137-144.
doi: 10.1016/j.tplants.2007.12.005 URL |
| [9] |
Hao YW, Wang XY, Li X, et al. Genome-wide identification, phylogenetic analysis, expression profiling, and protein-protein interaction properties of TOPLESS gene family members in tomato[J]. J Exp Bot, 2014, 65(4):1013-1023.
doi: 10.1093/jxb/ert440 URL |
| [10] |
Li HY, Huang KF, Du HM, et al. Genome-wide analysis of Gro/Tup1 family corepressors and their responses to hormones and abiotic stresses in maize[J]. J Plant Biol, 2016, 59(6):603-615.
doi: 10.1007/s12374-016-0333-8 URL |
| [11] |
Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis[J]. Science, 2008, 319(5868):1384-1386.
doi: 10.1126/science.1151461 pmid: 18258861 |
| [12] |
Nguyen D, Rieu I, Mariani C, et al. How plants handle multiple stresses:hormonal interactions underlying responses to abiotic stress and insect herbivory[J]. Plant Mol Biol, 2016, 91(6):727-740.
doi: 10.1007/s11103-016-0481-8 pmid: 27095445 |
| [13] |
Pauwels L, Barbero GF, Geerinck J, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling[J]. Nature, 2010, 464(7289):788-791.
doi: 10.1038/nature08854 URL |
| [14] |
You Y, Zhai Q, An C, et al. LEUNIG_HOMOLOG mediates MYC2-dependent transcriptional activation in cooperation with the coactivators HAC1 and MED25[J]. Plant Cell, 2019, 31(9):2187-2205.
doi: 10.1105/tpc.19.00115 URL |
| [15] |
Causier B, Ashworth M, Guo W, et al. The TOPLESS interactome:a framework for gene repression in Arabidopsis[J]. Plant Physiol, 2012, 158(1):423-438.
doi: 10.1104/pp.111.186999 pmid: 22065421 |
| [16] |
Gonzalez D, Bowen AJ, Carroll TS, et al. The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14(SWP)and CDK8(HEN3)to repress transcription[J]. Mol Cell Biol, 2007, 27(15):5306-5315.
doi: 10.1128/MCB.01912-06 URL |
| [17] |
Zhu J, Jeong JC, Zhu Y, et al. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance[J]. PNAS, 2008, 105(12):4945-4950.
doi: 10.1073/pnas.0801029105 URL |
| [18] |
Zhu Z, Xu F, Zhang Y, et al. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor[J]. PNAS, 2010, 107(31):13960-13965.
doi: 10.1073/pnas.1002828107 URL |
| [19] |
Shrestha B, Guragain B, Sridhar VV. Involvement of co-repressor LUH and the adapter proteins SLK1 and SLK2 in the regulation of abiotic stress response genes in Arabidopsis[J]. BMC Plant Biol, 2014, 14:54.
doi: 10.1186/1471-2229-14-54 pmid: 24564815 |
| [20] | 芦梅. 中国茶叶产业发展的管理模式与经济学分析[J]. 福建茶叶, 2016, 38(9):99-100. |
| Lu M. Management mode and economic analysis of tea industry development in China[J]. Tea Fujian, 2016, 38(9):99-100. | |
| [21] |
Liu SC, Jin JQ, Ma JQ, et al. Transcriptomic analysis of tea plant responding to drought stress and recovery[J]. PLoS One, 2016, 11(1):e0147306.
doi: 10.1371/journal.pone.0147306 URL |
| [22] |
Cheruiyot EK, Mumera LM, Ng’Etich WK, et al. High fertilizer rates increase susceptibility of tea to water stress[J]. J Plant Nutr, 2009, 33(1):115-129.
doi: 10.1080/01904160903392659 URL |
| [23] |
Wei CL, Yang H, Wang SB, et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality[J]. Proc Natl Acad Sci USA, 2018, 115(18):E4151-E4158.
doi: 10.1073/pnas.1719622115 URL |
| [24] |
Xia EH, Zhang HB, Sheng J, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynjournal[J]. Mol Plant, 2017, 10(6):866-877.
doi: 10.1016/j.molp.2017.04.002 URL |
| [25] |
Xia EH, Tong W, Hou Y, et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation[J]. Mol Plant, 2020, 13(7):1013-1026.
doi: 10.1016/j.molp.2020.04.010 URL |
| [26] |
Wang X, Feng H, Chang Y, et al. Population sequencing enhances understanding of tea plant evolution[J]. Nat Commun, 2020, 11(1):4447.
doi: 10.1038/s41467-020-18228-8 URL |
| [27] |
Zhang QJ, Li W, Li K, et al. The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons in driving genome size evolution[J]. Mol Plant, 2020, 13(7):935-938.
doi: 10.1016/j.molp.2020.04.009 URL |
| [28] | 项丽慧, 陈林, 余文权, 等. 茶树GH1基因家族鉴定及其在茶鲜叶萎凋过程的表达[J]. 应用与环境生物学报, 2020, 26(4):878-885. |
| Xiang LH, Chen L, Yu WQ, et al. Identification of the GH1 gene family in Camellia sinensis and expression analysis during the withering process of fresh tea leaves[J]. Chin J Appl Environ Biol, 2020, 26(4):878-885. | |
| [29] | 叶一隽, 李佳敏, 曹红利, 等. 茶树CsBBX基因家族的鉴定与表达[J]. 应用与环境生物学报, 2020, 26(6):1508-1516. |
| Ye YJ, Li JM, Cao HL, et al. Identification and expression analysis of the CsBBX gene family in tea plants[J]. Chin J Appl Environ Biol, 2020, 26(6):1508-1516. | |
| [30] |
Xia EH, Li FD, Tong W, et al. Tea Plant Information Archive:a comprehensive genomics and bioinformatics platform for tea plant[J]. Plant Biotechnol J, 2019, 17(10):1938-1953.
doi: 10.1111/pbi.v17.10 URL |
| [31] |
Zhou CZ, Zhu C, Fu H, et al. Genome-wide investigation of superoxide dismutase(SOD)gene family and their regulatory miRNAs reveal the involvement in abiotic stress and hormone response in tea plant(Camellia sinensis)[J]. PLoS One, 2019, 14(10):e0223609.
doi: 10.1371/journal.pone.0223609 URL |
| [32] |
Ouyang S, Zhu W, Hamilton J, et al. The TIGR Rice Genome Annotation Resource:improvements and new features[J]. Nucleic Acids Res, 2007, 35(database issue):D883-D887.
doi: 10.1093/nar/gkl976 URL |
| [33] | 马洪磊. 植物辅抑制因子TPL/TPR蛋白结构与EAR基序相互作用分子机理研究[D]. 上海:中国科学院上海药物研究所, 2016. |
| Ma HL. Structure of plant co-repressor TPL/TPR protein provides insights into mechanism of EAR motif binding[D]. Shanghai:Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 2016. | |
| [34] |
Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation[J]. Nat Rev Immunol, 2010, 10(1):24-35.
doi: 10.1038/nri2685 pmid: 20029446 |
| [35] |
Stahle MI, Kuehlich J, Staron L, et al. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis[J]. Plant Cell, 2009, 21(10):3105-3118.
doi: 10.1105/tpc.109.070458 URL |
| [36] |
Thines B, Katsir L, Melotto M, et al. JAZ repressor proteins are targets of the SCF(COI1)complex during jasmonate signalling[J]. Nature, 2007, 448(7154):661-665.
doi: 10.1038/nature05960 URL |
| [37] |
Chini A, Fonseca S, Fernández G, et al. The JAZ family of repressors is the missing link in jasmonate signalling[J]. Nature, 2007, 448(7154):666-671.
doi: 10.1038/nature06006 URL |
| [38] | 彭蕾. DNA序列的Exon-Intron结构的研究[D]. 北京:北京工业大学, 2001. |
| Peng L. Research of exon-intron structures in the DNA sequences[D]. Beijing:Beijing University of Technology, 2001. | |
| [39] | 王晓斌, 刘国仰. 有关内含子功能研究的新进展[J]. 中华医学遗传学杂志, 2000(3):211-212 |
| Wang XB, Liu GY. New research progress on the function of intron[J]. Chin J Med Genet, 2000(3):211-212 | |
| [40] | 单耀军. 内含子与基因间隔序列长度及基因表达量间关系的研究[D]. 保定:河北大学, 2007. |
| Shan YJ. Investigating the relation of introns to the length of intergenic sequences and gene expression levels[D]. Baoding:Hebei University, 2007. | |
| [41] |
Lorenzo O, Chico JM, Sánchez-Serrano JJ, et al. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis[J]. Plant Cell, 2004, 16(7):1938-1950.
pmid: 15208388 |
| [1] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [2] | LI Yuan-hong, GUO Yu-hao, CAO Yan, ZHU Zhen-zhou, WANG Fei-fei. Research Progress in the Microalgal Growth and Accumulation of Target Products Regulated by Exogenous Phytohormone [J]. Biotechnology Bulletin, 2023, 39(6): 61-72. |
| [3] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [4] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| [5] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [6] | YANG Chun-hong, DONG Lu, CHEN Lin, SONG Li. Characterization of Soybean VAS1 Gene Family and Its Involvement in Lateral Root Development [J]. Biotechnology Bulletin, 2023, 39(3): 133-142. |
| [7] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [8] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [9] | XU Rui, ZHU Ying-fang. The Key Roles of Mediator Complex in Plant Responses to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 54-60. |
| [10] | CHEN Guang-xia, LI Xiu-jie, JIANG Xi-long, SHAN Lei, ZHANG Zhi-chang, LI Bo. Research Progress in Plant Small Signaling Peptides Involved in Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(11): 61-73. |
| [11] | HAN Fang-ying, HU Xin, WANG Nan-nan, XIE Yu-hong, WANG Xiao-yan, ZHU Qiang. Research Progress in Response of DREBs to Abiotic Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(11): 86-98. |
| [12] | SUN Yu-tong, LIU De-shuai, QI Xun, FENG Mei, HUANG Xu-zheng, YAO Wen-kong. Advances in Jasmonic Acid Regulating Plant Growth and Development as Well as Stress [J]. Biotechnology Bulletin, 2023, 39(11): 99-109. |
| [13] | GE Wen-dong, WANG Teng-hui, MA Tian-yi, FAN Zhen-yu, WANG Yu-shu. Genome-wide Identification of the PRX Gene Family in Cabbage(Brassica oleracea L. var. capitata)and Expression Analysis Under Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 252-260. |
| [14] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [15] | AN Chang, LU Lin, SHEN Meng-qian, CHEN Sheng-zhen, YE Kang-zhuo, QIN Yuan, ZHENG Ping. Research Progress of bHLH Gene Family in Plants and Its Application Prospects in Medical Plants [J]. Biotechnology Bulletin, 2023, 39(10): 1-16. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||