








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (4): 153-162.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0915
Previous Articles Next Articles
ZU Guo-qiang( ), HU Zhe, WANG Qi, LI Guang-zhe, HAO Lin(
), HU Zhe, WANG Qi, LI Guang-zhe, HAO Lin( )
)
Received:2021-07-16
Online:2022-04-26
Published:2022-05-06
Contact:
HAO Lin
E-mail:1875224329@qq.com;haolinwj2001@163.com
ZU Guo-qiang, HU Zhe, WANG Qi, LI Guang-zhe, HAO Lin. Regulatory Role of Burkholderia sp. GD17 in Rice Seedling’s Responses to Cadmium Stress[J]. Biotechnology Bulletin, 2022, 38(4): 153-162.
| 基因 Gene | 基因ID Gene ID | 功能 Function | 引物序列 Primer sequence(5'-3') | 产物长度 Length/bp |
|---|---|---|---|---|
| ABCG5 | Os03g0281900 | 介导Cd进入液泡 Transporter pumping Cd into vacuole | F:AGGAGGACCTTCTGTACCCG R:CAGCATGATCGGGTTGTGGA | 245 |
| HMA1 | Os06g0690700 | 介导Cd进入液泡 Transporter pumping Cd into vacuole | F:CCGTAGCCTTCCCACTTGTC R:ACCGCCCTCCAATGAGTTTC | 140 |
| HMA3 | Os07g0232900 | 介导Cd进入液泡 Transporter pumping Cd into vacuole | F:CTGGCTCTGGTGATGCTTGT R:CCCAACCAGATGGAACGAGT | 219 |
| MT1a | Os11g0704500 | Cd络合和活性氧清除 Cd chelation and ROS scavenging | F:TGCGGAAAGAAGTACCCTGAC R:CCTCAAACTGCTGCGCCTTC | 100 |
| MT1c | Os12g0571100 | Cd络合和活性氧清除 Cd chelation and ROS scavenging | F:ATGTCGTGCGGTGGAAGTT R:TTGCAGTAGTGGTGGTGGTG | 109 |
| NRAMP5 | Os07g0257200 | 调节Cd的根-茎转运 Regulating Cd root-shoot transport | F:GGCCTCCAAAAATACGGGGT R:GCAAGAACAGATTGTGGGGC | 217 |
| PCS1 | Os05g0415200 | 介导络合素-Cd复合物的形成 Mediating the formation of phytochelatin -Cd complex | F:TGGATGTCGCTCGCTTCAAA R:CATGAACCCCCTGAGAAGCC | 107 |
| PCS2 | Os06g0102300 | 介导络合素-Cd复合物的形成 Mediating the formation of phytochelatin -Cd complex | F:GCAACTGGTCTACTCAGGGG R:GCTTTCATCTCTGCAACTCTGC | 101 |
Table 1 Primer sequences used for the analysis of gene expression
| 基因 Gene | 基因ID Gene ID | 功能 Function | 引物序列 Primer sequence(5'-3') | 产物长度 Length/bp |
|---|---|---|---|---|
| ABCG5 | Os03g0281900 | 介导Cd进入液泡 Transporter pumping Cd into vacuole | F:AGGAGGACCTTCTGTACCCG R:CAGCATGATCGGGTTGTGGA | 245 |
| HMA1 | Os06g0690700 | 介导Cd进入液泡 Transporter pumping Cd into vacuole | F:CCGTAGCCTTCCCACTTGTC R:ACCGCCCTCCAATGAGTTTC | 140 |
| HMA3 | Os07g0232900 | 介导Cd进入液泡 Transporter pumping Cd into vacuole | F:CTGGCTCTGGTGATGCTTGT R:CCCAACCAGATGGAACGAGT | 219 |
| MT1a | Os11g0704500 | Cd络合和活性氧清除 Cd chelation and ROS scavenging | F:TGCGGAAAGAAGTACCCTGAC R:CCTCAAACTGCTGCGCCTTC | 100 |
| MT1c | Os12g0571100 | Cd络合和活性氧清除 Cd chelation and ROS scavenging | F:ATGTCGTGCGGTGGAAGTT R:TTGCAGTAGTGGTGGTGGTG | 109 |
| NRAMP5 | Os07g0257200 | 调节Cd的根-茎转运 Regulating Cd root-shoot transport | F:GGCCTCCAAAAATACGGGGT R:GCAAGAACAGATTGTGGGGC | 217 |
| PCS1 | Os05g0415200 | 介导络合素-Cd复合物的形成 Mediating the formation of phytochelatin -Cd complex | F:TGGATGTCGCTCGCTTCAAA R:CATGAACCCCCTGAGAAGCC | 107 |
| PCS2 | Os06g0102300 | 介导络合素-Cd复合物的形成 Mediating the formation of phytochelatin -Cd complex | F:GCAACTGGTCTACTCAGGGG R:GCTTTCATCTCTGCAACTCTGC | 101 |
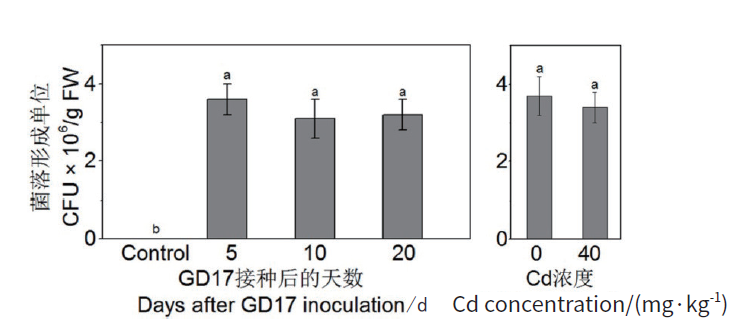
Fig. 1 Colonization efficiency of GD17 inside roots as indicated by colony-forming units(CFU) GD17 inoculation was performed during seed germination,CK was replaced with distilled water. The data were collected from three replicated experiments(n = 3)with 5 plants used in each batch of experiment,and represented as means ± SD. Bars with different lower-case letters indicate significant differences at P<0.05. The same below

Fig. 2 Effects of GD17 and(or)Cd stress on plant grow-th and Cd contents Twenty-day-old plants were evaluated. The data were collected from three replicated experiments(n=3)with 20 plants used in each batch of experiment,and represented as means ± SD. A and B:Fresh and dry weight of shoots. C and D:Fresh and dry weight of roots. E:Representative pictures showing the morphology of aboveground part. F and G:Cd contents in roots and shoots
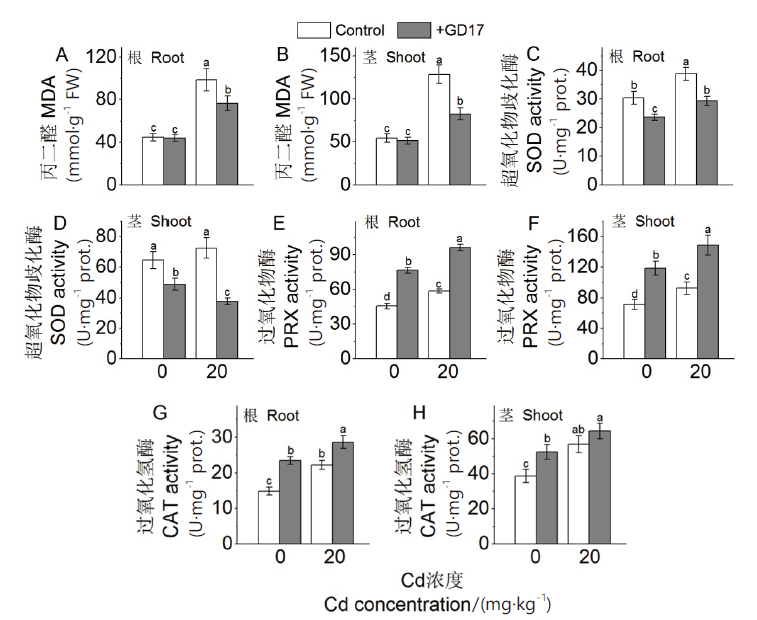
Fig. 3 Effects of GD17 and(or)Cd stress on malondialde-hyde(MDA)content of plants,and activities of antioxidation enzymes A and B:Malondialdehyde contents in roots and shoots. C and D:Superoxide dismutase activity in roots and shoots. E and F:Peroxidase activity in roots and shoots. G and H:Catalase activity in roots and shoots
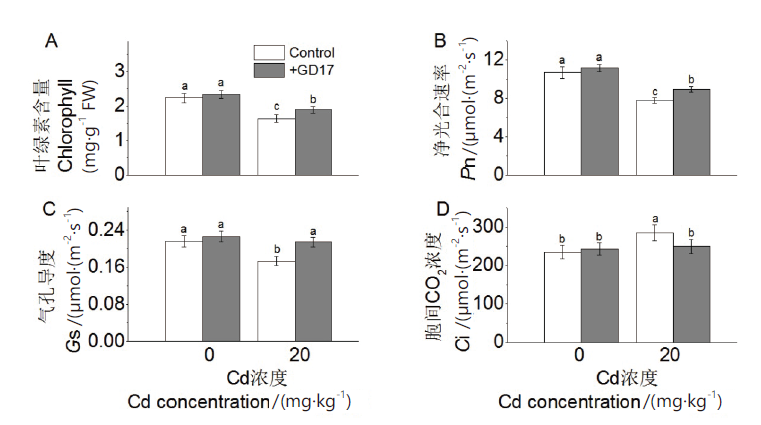
Fig. 4 Effects of GD17 and(or)Cd stress on photosynth-esis-related parameters A:Total chlorophyll content. B:Photosynthetic rate(Pn). C:Stomatal conductance(Gs). D:Intercellular CO2 concentration(Ci)

Fig. 5 Effects of GD17 and(or)Cd stress on chlorophyll fluorescence parameters Fv/Fm:Maximal photochemical efficiency of PSII. ФPSII:Actual photochemical efficiency of PSII. ETR:Electron transport rate. NPQ:Non-photochemical quenching of fluorescence
| [1] |
Fagerberg B, Barregard L, Sallsten G, et al. Cadmium exposure and atherosclerotic carotid plaques——results from the Malmö diet and cancer study[J]. Environ Res, 2015, 136:67-74.
doi: 10.1016/j.envres.2014.11.004 pmid: 25460622 |
| [2] |
Fasani E, Manara A, Martini F, et al. The potential of genetic engineering of plants for the remediation of soils contaminated with heavy metals[J]. Plant Cell Environ, 2018, 41(5):1201-1232.
doi: 10.1111/pce.12963 URL |
| [3] |
Wang P, Chen H, Kopittke PM, et al. Cadmium contamination in agricultural soils of China and the impact on food safety[J]. Environ Pollut, 2019, 249:1038-1048.
doi: 10.1016/j.envpol.2019.03.063 URL |
| [4] | Qadir S, Jamshieed S, Rasool S, et al. Modulation of plant growth and metabolism in cadmium-enriched environments[M]//Reviews of environmental contamination and toxicology. Whitacre DM. Switzerland: Springer International Publishing, 2014: 51-88. |
| [5] |
Huybrechts M, Cuypers A, Deckers J, et al. Cadmium and plant development:an agony from seed to seed[J]. Int J Mol Sci, 2019, 20(16):3971.
doi: 10.3390/ijms20163971 URL |
| [6] |
Sui FQ, Chang JD, Tang Z, et al. Nramp5 expression and functionality likely explain higher cadmium uptake in rice than in wheat and maize[J]. Plant Soil, 2018, 433(1/2):377-389.
doi: 10.1007/s11104-018-3849-5 URL |
| [7] |
Zhao FJ, Wang P. Arsenic and cadmium accumulation in rice and mitigation strategies[J]. Plant Soil, 2020, 446(1):1-21.
doi: 10.1007/s11104-019-04374-6 URL |
| [8] | Song Y, Wang Y, Mao W, et al. Dietary cadmium exposure assessment among the Chinese population[J]. PLoS One, 2017, 12(5):e0177978. |
| [9] |
Rizwan M, Ali S, Adrees M, et al. Cadmium stress in rice:toxic effects, tolerance mechanisms, and management:a critical review[J]. Environ Sci Pollut Res Int, 2016, 23(18):17859-17879.
doi: 10.1007/s11356-016-6436-4 URL |
| [10] |
Pramanik K, Mitra S, Sarkar A, et al. Characterization of cadmium-resistant Klebsiella pneumoniae MCC 3091 promoted rice seedling growth by alleviating phytotoxicity of cadmium[J]. Environ Sci Pollut Res Int, 2017, 24(31):24419-24437.
doi: 10.1007/s11356-017-0033-z URL |
| [11] |
郭英, 杨萍, 张丹雨, 等. 野大豆多功能根际促生菌的筛选鉴定和促生效果研究[J]. 生物技术通报, 2018, 34(10):108-115.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-0437 |
| Guo Y, Yang P, Zhang DY, et al. Screening, identification and growth-promoting effect of multifunction rhizosphere growth-promoting strain of wild soybean[J]. Biotechnol Bull, 2018, 34(10):108-115. | |
| [12] |
Yang AZ, Akhtar SS, Fu Q, et al. Burkholderia phytofirmans PsJN stimulate growth and yield of quinoa under salinity stress[J]. Plants, 2020, 9(6):672.
doi: 10.3390/plants9060672 URL |
| [13] | 李合生. 植物生理生化实验原理和技术[M]. 北京: 高等教育出版社, 2000. |
| Li HS. Principles and techniques of plant physiological biochemical experiment[M]. Beijing: Higher Education Press, 2000. | |
| [14] | 王学奎, 黄见良. 植物生理生化实验原理与技术[M]. 3版. 北京: 高等教育出版社, 2015. |
| Wang XK, Huang JL. Principles and techniques of plant physiological biochemical experiment[M]. 3rd ed. Beijing: Higher Education Press, 2015. | |
| [15] |
Hemeda HM, Klein BP. Effects of naturally occurring antioxidants on peroxidase activity of vegetable extracts[J]. J Food Sci, 1990, 55(1):184-185.
doi: 10.1111/j.1365-2621.1990.tb06048.x URL |
| [16] |
Wang YY, Wang Y, Li GZ, et al. Salicylic acid-altering Arabidopsis plant response to cadmium exposure:Underlying mechanisms affecting antioxidation and photosynjournal-related processes[J]. Ecotoxicol Environ Saf, 2019, 169:645-653.
doi: 10.1016/j.ecoenv.2018.11.062 URL |
| [17] |
Narsai R, Ivanova A, Ng S, et al. Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets[J]. BMC Plant Biol, 2010, 10:56.
doi: 10.1186/1471-2229-10-56 pmid: 20353606 |
| [18] |
Zhang JH, Li Q, Zeng YF, et al. Bioaccumulation and distribution of cadmium by Burkholderia cepacia GYP1 under oligotrophic condition and mechanism analysis at proteome level[J]. Ecotoxicol Environ Saf, 2019, 176:162-169.
doi: 10.1016/j.ecoenv.2019.03.091 URL |
| [19] |
Yang ZH, Zhang Z, Chai LY, et al. Bioleaching remediation of heavy metal-contaminated soils using Burkholderia sp. Z-90[J]. J Hazard Mater, 2016, 301:145-152.
doi: 10.1016/j.jhazmat.2015.08.047 URL |
| [20] |
Wang X, Zhang X, Liu XM, et al. Physiological, biochemical and proteomic insight into integrated strategies of an endophytic bacterium Burkholderia cenocepacia strain YG-3 response to cadmium stress[J]. Metallomics, 2019, 11(7):1252-1264.
doi: 10.1039/c9mt00054b URL |
| [21] | Wang CR, Huang YC, Yang XR, et al. Burkholderia sp. Y4 inhibits cadmium accumulation in rice by increasing essential nutrient uptake and preferentially absorbing cadmium[J]. Chemosphere, 2020, 252:126603. |
| [22] |
Cuypers A, Plusquin M, Remans T, et al. Cadmium stress:an oxidative challenge[J]. Biometals, 2010, 23(5):927-940.
doi: 10.1007/s10534-010-9329-x pmid: 20361350 |
| [23] |
Sarkar A, Pramanik K, Mitra S, et al. Enhancement of growth and salt tolerance of rice seedlings by ACC deaminase-producing Burkholderia sp. MTCC 12259[J]. J Plant Physiol, 2018, 231:434-442.
doi: 10.1016/j.jplph.2018.10.010 URL |
| [24] |
Mitra S, Pramanik K, Sarkar A, et al. Bioaccumulation of cadmium by Enterobacter sp. and enhancement of rice seedling growth under cadmium stress[J]. Ecotoxicol Environ Saf, 2018, 156:183-196.
doi: 10.1016/j.ecoenv.2018.03.001 URL |
| [25] | Liu HJ, Yang L, Li N, et al. Cadmium toxicity reduction in rice(Oryza sativa L.)through iron addition during primary reaction of photosynjournal[J]. Ecotoxicol Environ Saf, 2020, 200:110746. |
| [26] |
Khanna, Kohli, Ohri, et al. Microbial fortification improved photosynthetic efficiency and secondary metabolism in Lycopersicon esculentum plants under Cd stress[J]. Biomolecules, 2019, 9(10):581.
doi: 10.3390/biom9100581 URL |
| [27] |
Baker NR. Chlorophyll fluorescence:a probe of photosynjournal in vivo[J]. Annu Rev Plant Biol, 2008, 59:89-113.
doi: 10.1146/annurev.arplant.59.032607.092759 pmid: 18444897 |
| [28] |
Huang W, Zhang SB, Liu T. Moderate photoinhibition of photosystem II significantly affects linear electron flow in the shade-demanding plant Panax notoginseng[J]. Front Plant Sci, 2018, 9:637.
doi: 10.3389/fpls.2018.00637 pmid: 29868090 |
| [29] | Wu Y, Ma L, Liu Q, et al. Pseudomonas fluorescens promote photosynjournal, carbon fixation and cadmium phytoremediation of hyperaccumulator Sedum alfredii[J]. Sci Total Environ, 2020, 726:138554. |
| [30] |
Takahashi R, Bashir K, Ishimaru Y, et al. The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice[J]. Plant Signal Behav, 2012, 7(12):1605-1607.
doi: 10.4161/psb.22454 pmid: 23072989 |
| [31] |
Cong W, Miao Y, Xu L, et al. Transgenerational memory of gene expression changes induced by heavy metal stress in rice(Oryza sativa L.)[J]. BMC Plant Biol, 2019, 19(1):282.
doi: 10.1186/s12870-019-1887-7 URL |
| [32] |
Ueno D, Yamaji N, Kono I, et al. Gene limiting cadmium accumulation in rice[J]. PNAS, 2010, 107(38):16500-16505.
doi: 10.1073/pnas.1005396107 URL |
| [33] |
Ishikawa S, Ishimaru Y, Igura M, et al. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice[J]. PNAS, 2012, 109(47):19166-19171.
doi: 10.1073/pnas.1211132109 pmid: 23132948 |
| [34] |
Ishimaru Y, Takahashi R, Bashir K, et al. Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport[J]. Sci Rep, 2012, 2:286.
doi: 10.1038/srep00286 URL |
| [35] |
Chang JD, Huang S, Konishi N, et al. Overexpression of the manganese/cadmium transporter OsNRAMP5 reduces cadmium accumulation in rice grain[J]. J Exp Bot, 2020, 71(18):5705-5715.
doi: 10.1093/jxb/eraa287 URL |
| [36] |
Tang L, Mao B, Li Y, et al. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield[J]. Sci Rep, 2017, 7(1):14438.
doi: 10.1038/s41598-017-14832-9 pmid: 29089547 |
| [37] | Yamauchi T, Fukazawa A, Nakazono M. METALLOTHIONEIN genes encoding ROS scavenging enzymes are down-regulated in the root cortex during inducible aerenchyma formation in rice[J]. Plant Signal Behav, 2017, 12(11):e1388976. |
| [38] |
Kim YO, Kang H. Comparative expression analysis of genes encoding metallothioneins in response to heavy metals and abiotic stresses in rice(Oryza sativa)and Arabidopsis thaliana[J]. Biosci Biotechnol Biochem, 2018, 82(9):1656-1665.
doi: 10.1080/09168451.2018.1486177 URL |
| [39] | Yamazaki S, Ueda Y, Mukai A, et al. Rice phytochelatin synthases OsPCS1 and OsPCS2 make different contributions to cadmium and arsenic tolerance[J]. Plant Direct, 2018, 2(1):e00034. |
| [40] |
Uraguchi S, Tanaka N, Hofmann C, et al. Phytochelatin synthase has contrasting effects on cadmium and arsenic accumulation in rice grains[J]. Plant Cell Physiol, 2017, 58(10):1730-1742.
doi: 10.1093/pcp/pcx114 pmid: 29016913 |
| [41] |
Brunetti P, Zanella L, De Paolis A, et al. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis[J]. J Exp Bot, 2015, 66(13):3815-3829.
doi: 10.1093/jxb/erv185 URL |
| [42] |
Matsuda S, Funabiki A, Furukawa K, et al. Genome-wide analysis and expression profiling of half-size ABC protein subgroup G in rice in response to abiotic stress and phytohormone treatments[J]. Mol Genet Genomics, 2012, 287(10):819-835.
doi: 10.1007/s00438-012-0719-3 URL |
| [43] |
Fu S, Lu Y, Zhang X, et al. The ABC transporter ABCG36 is required for cadmium tolerance in rice[J]. J Exp Bot, 2019, 70(20):5909-5918.
doi: 10.1093/jxb/erz335 URL |
| [44] |
Oda K, Otani M, Uraguchi S, et al. Rice ABCG43 is Cd inducible and confers Cd tolerance on yeast[J]. Biosci Biotechnol Biochem, 2011, 75(6):1211-1213.
doi: 10.1271/bbb.110193 URL |
| [1] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [2] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [3] | KANG Ling-yun, HAN Lu-lu, HAN De-ping, CHEN Jian-sheng, GAN Han-ling, XING Kai, MA You-ji, CUI Kai. Effect of Melatonin on Protecting the Jejunum Mucosal Epithelial Cells from Oxidative Stress Damage [J]. Biotechnology Bulletin, 2023, 39(9): 291-299. |
| [4] | WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq [J]. Biotechnology Bulletin, 2023, 39(8): 173-184. |
| [5] | LIU Bao-cai, CHEN Jing-ying, ZHANG Wu-jun, HUANG Ying-zhen, ZHAO Yun-qing, LIU Jian-chao, WEI Zhi-cheng. Characteristics Analysis of Seed Microrhizome Gene Expression of Polygonatum cyrtonema [J]. Biotechnology Bulletin, 2023, 39(8): 220-233. |
| [6] | YAO Sha-sha, WANG Jing-jing, WANG Jun-jie, LIANG Wei-hong. Molecular Mechanisms of Rice Grain Size Regulation Related to Plant Hormone Signaling Pathways [J]. Biotechnology Bulletin, 2023, 39(8): 80-90. |
| [7] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [8] | YU Hui, WANG Jing, LIANG Xin-xin, XIN Ya-ping, ZHOU Jun, ZHAO Hui-jun. Isolation and Functional Verification of Genes Responding to Iron and Cadmium Stresses in Lycium barbarum [J]. Biotechnology Bulletin, 2023, 39(7): 195-205. |
| [9] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [10] | LIANG Cheng-gang, WANG Yan, LI Tian, OHSUGI Ryu, AOKI Naohiro. Effect of SP1 on Panicle Architecture by Regulating Carbohydrate Remobilization [J]. Biotechnology Bulletin, 2023, 39(5): 152-159. |
| [11] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [12] | ZHOU Ding-ding, LI Hui-hu, TANG Xing-yong, YU Fa-xin, KONG Dan-yu, LIU Yi. Research Progress in the Biosynthesis and Regulation of Glycyrrhizic Acid and Liquiritin [J]. Biotechnology Bulletin, 2023, 39(5): 44-53. |
| [13] | WANG Qi, HU Zhe, FU Wei, LI Guang-zhe, HAO Lin. Regulation of Burkholderia sp. GD17 on the Drought Tolerance of Cucumber Seedlings [J]. Biotechnology Bulletin, 2023, 39(3): 163-175. |
| [14] | YANG Mao, LIN Yu-feng, DAI Yang-shuo, PAN Su-jun, PENG Wei-ye, YAN Ming-xiong, LI Wei, WANG Bing, DAI Liang-ying. OsDIS1 Negatively Regulates Rice Drought Tolerance Through Antioxidant Pathways [J]. Biotechnology Bulletin, 2023, 39(2): 88-95. |
| [15] | JIANG Min-xuan, LI Kang, LUO Liang, LIU Jian-xiang, LU Hai-ping. Advances on the Expressions of Foreign Proteins in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 110-122. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||