








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (12): 214-222.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0290
Previous Articles Next Articles
WANG Shuai1,2( ), YUAN Kun2, HE Qi-guang2, HU Yi-yu2, FENG Cheng-tian2, WANG Zhen-hui2, LIU Jin-ping1, LIU Hui2(
), YUAN Kun2, HE Qi-guang2, HU Yi-yu2, FENG Cheng-tian2, WANG Zhen-hui2, LIU Jin-ping1, LIU Hui2( )
)
Received:2022-03-30
Online:2022-12-26
Published:2022-12-29
Contact:
LIU Hui
E-mail:791647230@qq.com;liuhui@catas.cn
WANG Shuai, YUAN Kun, HE Qi-guang, HU Yi-yu, FENG Cheng-tian, WANG Zhen-hui, LIU Jin-ping, LIU Hui. Cloning and Expression Analysis of HbCXXS1,a Thioredoxin Gene in Hevea brasiliensis[J]. Biotechnology Bulletin, 2022, 38(12): 214-222.
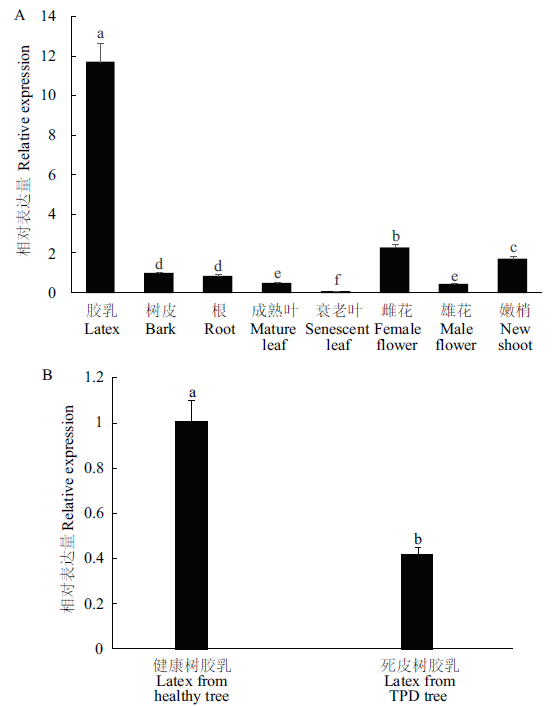
Fig. 4 Expressions of HbCXXS1 in various tissues(A)and latex from healthy and TPD rubber tree(B) Different lowercase letters indicate a significant difference(P<0.05). The same below
| [1] |
Serrato AJ, Rojas-González JA, Torres-Romero D, et al. Thioredoxins m are major players in the multifaceted light-adaptive response in Arabidopsis thaliana[J]. Plant J, 2021, 108(1):120-133.
doi: 10.1111/tpj.15429 URL |
| [2] |
Bohrer AS, Massot V, Innocenti G, et al. New insights into the reduction systems of plastidial thioredoxins point out the unique properties of thioredoxin z from Arabidopsis[J]. J Exp Bot, 2012, 63(18):6315-6323.
doi: 10.1093/jxb/ers283 URL |
| [3] | 孙虎, 薛保国, 杨丽荣, 等. 植物硫氧还蛋白系统[J]. 基因组学与应用生物学, 2010, 29(4):748-753. |
| Sun H, Xue BG, Yang LR, et al. Plant thioredoxin system[J]. Genom Appl Biol, 2010, 29(4):748-753. | |
| [4] |
Collet JF, Messens J. Structure, function, and mechanism of thioredoxin proteins[J]. Antioxid Redox Signal, 2010, 13(8):1205-1216.
doi: 10.1089/ars.2010.3114 URL |
| [5] |
Gelhaye E, Rouhier N, Navrot N, et al. The plant thioredoxin system[J]. Cell Mol Life Sci, 2005, 62(1):24-35.
pmid: 15619004 |
| [6] |
Meyer Y, Belin C, Delorme-Hinoux V, et al. Thioredoxin and glutaredoxin systems in plants:molecular mechanisms, crosstalks, and functional significance[J]. Antioxid Redox Signal, 2012, 17(8):1124-1160.
doi: 10.1089/ars.2011.4327 URL |
| [7] |
Geigenberger P, Thormählen I, Daloso DM, et al. The unprecedented versatility of the plant thioredoxin system[J]. Trends Plant Sci, 2017, 22(3):249-262.
doi: S1360-1385(16)30221-7 pmid: 28139457 |
| [8] |
Gelhaye E, Rouhier N, Jacquot JP. The thioredoxin h system of higher plants[J]. Plant Physiol Biochem, 2004, 42(4):265-271.
doi: 10.1016/j.plaphy.2004.03.002 URL |
| [9] |
Chibani K, Wingsle G, Jacquot JP, et al. Comparative genomic study of the thioredoxin family in photosynthetic organisms with emphasis on Populus trichocarpa[J]. Mol Plant, 2009, 2(2):308-322.
doi: 10.1093/mp/ssn076 pmid: 19825616 |
| [10] | Meyer Y, Vignols F, Reichheld JP. Classification of plant thioredoxins by sequence similarity and intron position[J]. Methods Enzymol, 2002, 347:394-402. |
| [11] |
Serrato AJ, Guilleminot J, Meyer Y, et al. AtCXXS:atypical members of the Arabidopsis thaliana thioredoxin h family with a remarkably high disulfide isomerase activity[J]. Physiol Plant, 2008, 133(3):611-622.
doi: 10.1111/j.1399-3054.2008.01093.x URL |
| [12] |
Gelhaye E, Rouhier N, Jacquot JP. Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction[J]. FEBS Lett, 2003, 555(3):443-448.
pmid: 14675753 |
| [13] |
Renard M, Alkhalfioui F, Schmitt-Keichinger C, et al. Identification and characterization of thioredoxin h isoforms differentially expressed in germinating seeds of the model legume Medicago truncatula[J]. Plant Physiol, 2011, 155(3):1113-1126.
doi: 10.1104/pp.110.170712 pmid: 21239621 |
| [14] |
Ancín M, Fernández-San Millán A, Larraya L, et al. Overexpression of thioredoxin m in tobacco chloroplasts inhibits the protein kinase STN7 and alters photosynthetic performance[J]. J Exp Bot, 2018, 70(3):1005-1016.
doi: 10.1093/jxb/ery415 URL |
| [15] |
Richter AS, Pérez-Ruiz JM, Cejudo FJ, et al. Redox-control of chlorophyll biosynthesis mainly depends on thioredoxins[J]. FEBS Lett, 2018, 592(18):3111-3115.
doi: 10.1002/1873-3468.13216 pmid: 30076598 |
| [16] |
Zhang H, Zhang TT, Liu H, et al. Thioredoxin-mediated ROS homeostasis explains natural variation in plant regeneration[J]. Plant Physiol, 2018, 176(3):2231-2250.
doi: 10.1104/pp.17.00633 pmid: 28724620 |
| [17] |
Ying YH, Yue WH, Wang SD, et al. Two h-type thioredoxins interact with the E2 ubiquitin conjugase PHO2 to fine-tune phosphate homeostasis in rice[J]. Plant Physiol, 2017, 173(1):812-824.
doi: 10.1104/pp.16.01639 URL |
| [18] |
Bashandy T, Guilleminot J, Vernoux T, et al. Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling[J]. Plant Cell, 2010, 22(2):376-391.
doi: 10.1105/tpc.109.071225 URL |
| [19] |
Luan JY, Dong JX, Song X, et al. Overexpression of Tamarix hispida ThTrx5 confers salt tolerance to Arabidopsis by activating stress response signals[J]. Int J Mol Sci, 2020, 21(3):1165.
doi: 10.3390/ijms21031165 URL |
| [20] |
Zhang CJ, Zhao BC, Ge WN, et al. An apoplastic h-type thioredoxin is involved in the stress response through regulation of the apoplastic reactive oxygen species in rice[J]. Plant Physiol, 2011, 157(4):1884-1899.
doi: 10.1104/pp.111.182808 URL |
| [21] |
Mata-Pérez C, Spoel SH. Thioredoxin-mediated redox signalling in plant immunity[J]. Plant Sci, 2019, 279:27-33.
doi: S0168-9452(17)31251-7 pmid: 30709489 |
| [22] |
Laloi C, Mestres-Ortega D, Marco Y, et al. The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor[J]. Plant Physiol, 2004, 134(3):1006-1016.
pmid: 14976236 |
| [23] |
Torres-Rodríguez MD, Cruz-Zamora Y, Juárez-Díaz JA, et al. NaTrxh is an essential protein for pollen rejection in Nicotiana by increasing S-RNase activity[J]. Plant J, 2020, 103(4):1304-1317.
doi: 10.1111/tpj.14802 URL |
| [24] | 郭晋隆, 郑蕊, 凌辉, 等. 甘蔗硫氧还蛋白基因ScTRXh1的克隆及表达特性分析[J]. 基因组学与应用生物学, 2012, 31(6):574-581. |
| Guo JL, Zheng R, Ling H, et al. Cloning and expression analysis of a thioredoxin gene ScTRXh1 from sugarcane(Saccharum complex)[J]. Genom Appl Biol, 2012, 31(6):574-581. | |
| [25] | 李霞, 潘春柳, 苏桂军, 等. 花生硫氧还蛋白AhTRX h基因的克隆及表达分析[J]. 热带作物学报, 2021, 42(5):1252-1260. |
| Li X, Pan CL, Su GJ, et al. Cloning and expression analysis of thioredoxin gene AhTRX h in peanut(Arachis hypogaea)[J]. Chin J Trop Crops, 2021, 42(5):1252-1260. | |
| [26] | 李梦园, 樊亚栋, 张新宁, 等. 小麦TaTrxh9基因的序列特征及其对渗透胁迫的响应[J]. 西北植物学报, 2019, 39(5):896-903. |
| Li MY, Fan YD, Zhang XN, et al. Sequence characteristics of TaTrxh9 gene and its response to osmotic stresses in wheat[J]. Acta Bot Boreali Occidentalia Sin, 2019, 39(5):896-903. | |
| [27] |
Li HL, Lu HZ, Guo D, et al. Molecular characterization of a thioredoxin h gene(HbTRX1)from Hevea brasiliensis showing differential expression in latex between self-rooting juvenile clones and donor clones[J]. Mol Biol Rep, 2011, 38(3):1989-1994.
doi: 10.1007/s11033-010-0321-x URL |
| [28] | Montoro P, Wu S, Favreau B, et al. Transcriptome analysis in Hevea brasiliensis latex revealed changes in hormone signalling pathways during ethephon stimulation and consequent Tapping Panel Dryness[J]. Sci Reports, 2018, 8:8483. |
| [29] |
李双江, 冯成天, 胡义钰, 等. 橡胶树HbDHAR2基因克隆及表达分析[J]. 华北农学报, 2021, 36(3):25-32.
doi: 10.7668/hbnxb.20191875 |
| Li SJ, Feng CT, Hu YY, et al. Cloning and expression analysis of HbDHAR2 gene from Hevea brasiliensis[J]. Acta Agric Boreali Sin, 2021, 36(3):25-32. | |
| [30] |
Nuruzzaman M, Gupta M, Zhang CJ, et al. Sequence and expression analysis of the thioredoxin protein gene family in rice[J]. Mol Genet Genomics, 2008, 280(2):139-151.
doi: 10.1007/s00438-008-0351-4 pmid: 18491141 |
| [31] |
Balsera M, Buchanan BB. Evolution of the thioredoxin system as a step enabling adaptation to oxidative stress[J]. Free Radic Biol Med, 2019, 140:28-35.
doi: 10.1016/j.freeradbiomed.2019.03.003 URL |
| [32] |
Zhang Y, Leclercq J, Montoro P. Reactive oxygen species in Hevea brasiliensis latex and relevance to Tapping Panel Dryness[J]. Tree Physiol, 2017, 37(2):261-269.
doi: 10.1093/treephys/tpw106 pmid: 27903918 |
| [1] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [2] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [3] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [4] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [5] | LI Yuan-hong, GUO Yu-hao, CAO Yan, ZHU Zhen-zhou, WANG Fei-fei. Research Progress in the Microalgal Growth and Accumulation of Target Products Regulated by Exogenous Phytohormone [J]. Biotechnology Bulletin, 2023, 39(6): 61-72. |
| [6] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [7] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [8] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| [9] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [10] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [11] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [12] | YANG Jun-zhao, ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei. Structure and Function Analysis of Novel GH5 Multi-domain Cellulase [J]. Biotechnology Bulletin, 2023, 39(4): 71-80. |
| [13] | YANG Chun-hong, DONG Lu, CHEN Lin, SONG Li. Characterization of Soybean VAS1 Gene Family and Its Involvement in Lateral Root Development [J]. Biotechnology Bulletin, 2023, 39(3): 133-142. |
| [14] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [15] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||