








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (12): 204-213.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0330
Previous Articles Next Articles
SUN Yan1,2( ), WANG Jin-gang1(
), WANG Jin-gang1( ), ZANG Dan-dan2(
), ZANG Dan-dan2( ), ZHAO Heng-tian2, LIU Shu-hua2
), ZHAO Heng-tian2, LIU Shu-hua2
Received:2022-03-19
Online:2022-12-26
Published:2022-12-29
Contact:
WANG Jin-gang,ZANG Dan-dan
E-mail:yansuns@163.com;wangjingang99@neau.edu.cn;zangdandan@iga.ac.cn
SUN Yan, WANG Jin-gang, ZANG Dan-dan, ZHAO Heng-tian, LIU Shu-hua. Transcriptome Analysis of Lonicera caerulea Fruits at Different Developmental Stages[J]. Biotechnology Bulletin, 2022, 38(12): 204-213.
| 基因Gene | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|
| OM964861 | CCCTGGGGTC | CTCAGAGGAATTGAATTC |
| OM964862 | GCAAGAACTATTTAACAG | CTCACCACCAGTTTG |
| OM964867 | GTCGAATGTGTCAAC | CATTAGATCCTGGCTGC |
| OM964864 | CGAGCTAATGGACACGGTG | GTAGCCTTTCTCTAATCC |
| OM964868 | CTATTCCGAGAGATTTG | GTTCTTTGGTTCCCATC |
| OM964869 | GGTTTACATTTTAGCGAC | CAACTTGTCCTCACCAAC |
| OM964870 | CTTTGAATCCCATTGATTC | GAATTGCTACTGGCAAGC |
| OM964871 | GCTGGTCATTTTACAG | CACTGCCCTTATTAGTG |
| OM964872 | CCACCGACCTTCCC | GACATGTTCCGCGCTC |
| OM964873 | CAACGATTTCCGAAC | CCGGTGTCACCTTTTCC |
| MT344113 | CGTCGTTTGTGACAATGG | GGTCCCACACATGACCC |
| MT344114 | GTCTGTGCCGAACACGGC | CAGTATAATGGCCTTTAGC |
Table 1 Primer sequences used in RT-qPCR validation of transcriptome data
| 基因Gene | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|
| OM964861 | CCCTGGGGTC | CTCAGAGGAATTGAATTC |
| OM964862 | GCAAGAACTATTTAACAG | CTCACCACCAGTTTG |
| OM964867 | GTCGAATGTGTCAAC | CATTAGATCCTGGCTGC |
| OM964864 | CGAGCTAATGGACACGGTG | GTAGCCTTTCTCTAATCC |
| OM964868 | CTATTCCGAGAGATTTG | GTTCTTTGGTTCCCATC |
| OM964869 | GGTTTACATTTTAGCGAC | CAACTTGTCCTCACCAAC |
| OM964870 | CTTTGAATCCCATTGATTC | GAATTGCTACTGGCAAGC |
| OM964871 | GCTGGTCATTTTACAG | CACTGCCCTTATTAGTG |
| OM964872 | CCACCGACCTTCCC | GACATGTTCCGCGCTC |
| OM964873 | CAACGATTTCCGAAC | CCGGTGTCACCTTTTCC |
| MT344113 | CGTCGTTTGTGACAATGG | GGTCCCACACATGACCC |
| MT344114 | GTCTGTGCCGAACACGGC | CAGTATAATGGCCTTTAGC |
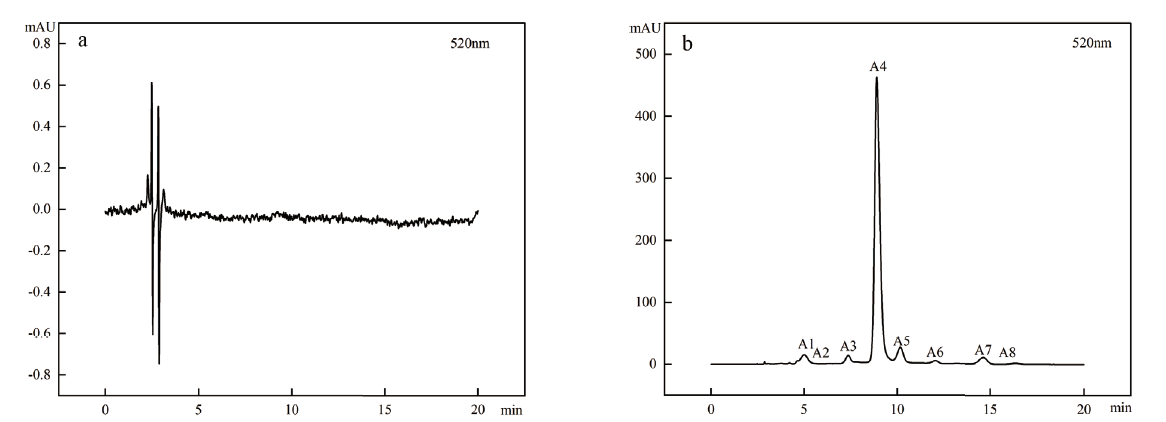
Fig. 2 HPLC of ‘Zhongkelan No.1’ fruit at different developmental stages a:Green ripening stage of Lonicera caerulea fruit. b:Ripening stage of Lonicera caerulea fruit. A1:Cyanidin-3,5-diglucoside. A2:Delphinidin-3-O-glucoside. A3:Delphinidin-3-O-rutinosidchlorid. A4:Cyanidin-3-O-glucoside. A5:Cyanidin-3-O-rutinoside. A6:Pelargonidin-3-O-glucoside. A7:Peonidin-3-O-glucoside. A8:Peonidin-3-O-rutinoside
| Sample | Clean reads | Clean bases | GC Content/% | Q30/% |
|---|---|---|---|---|
| B01 | 23 970 275 | 7 147 808 408 | 46.32 | 94.50 |
| B02 | 21 515 272 | 6 404 727 658 | 47.02 | 94.18 |
| B03 | 21 364 950 | 6 366 085 640 | 46.64 | 93.89 |
| G01 | 20 399 412 | 6 074 481 494 | 49.24 | 94.10 |
| G02 | 22 320 706 | 6 652 355 802 | 48.67 | 94.26 |
| G03 | 25 781 714 | 7 672 240 582 | 49.38 | 93.90 |
Table 3 Statistics of sequencing data
| Sample | Clean reads | Clean bases | GC Content/% | Q30/% |
|---|---|---|---|---|
| B01 | 23 970 275 | 7 147 808 408 | 46.32 | 94.50 |
| B02 | 21 515 272 | 6 404 727 658 | 47.02 | 94.18 |
| B03 | 21 364 950 | 6 366 085 640 | 46.64 | 93.89 |
| G01 | 20 399 412 | 6 074 481 494 | 49.24 | 94.10 |
| G02 | 22 320 706 | 6 652 355 802 | 48.67 | 94.26 |
| G03 | 25 781 714 | 7 672 240 582 | 49.38 | 93.90 |
| 不同长度区间 Length range/bp | 转录本序列数量 Transcripts | 单基因序列数量 Unigene |
|---|---|---|
| 200-300 | 37 818(16.02%) | 29 441(36.00%) |
| 300-500 | 34 253(14.51%) | 20 582(25.17%) |
| 500-1 000 | 44 975(19.05%) | 14 415(17.63%) |
| 1 000-2 000 | 60 698(25.72%) | 9 656(11.81%) |
| 2 000+ | 58 289(24.70%) | 7 683(9.40%) |
| Total number | 236 033 | 81 777 |
| Total Length | 323 833 751 | 62 424 481 |
| N50 Length | 2 186 | 1 421 |
| Mean Length | 1 371.99 | 763.35 |
Table 4 Assembly result statistics
| 不同长度区间 Length range/bp | 转录本序列数量 Transcripts | 单基因序列数量 Unigene |
|---|---|---|
| 200-300 | 37 818(16.02%) | 29 441(36.00%) |
| 300-500 | 34 253(14.51%) | 20 582(25.17%) |
| 500-1 000 | 44 975(19.05%) | 14 415(17.63%) |
| 1 000-2 000 | 60 698(25.72%) | 9 656(11.81%) |
| 2 000+ | 58 289(24.70%) | 7 683(9.40%) |
| Total number | 236 033 | 81 777 |
| Total Length | 323 833 751 | 62 424 481 |
| N50 Length | 2 186 | 1 421 |
| Mean Length | 1 371.99 | 763.35 |
| 数据库 Database | 注释的单基因数Number of annotated unigenes | 300 bp≤长度<1 000 bp 300 bp≤Length<1 000 bp | 长度≥1 000 bp Length≥1 000 bp |
|---|---|---|---|
| COG | 8 056 | 1 670 | 5 646 |
| GO | 17 733 | 5 300 | 9 680 |
| KEGG | 9 814 | 2 805 | 5 673 |
| KOG | 15 668 | 4 567 | 8 845 |
| Pfam | 18 933 | 5 043 | 12 068 |
| Swissprot | 19 579 | 5 863 | 11 158 |
| eggNOG | 26 275 | 8 298 | 14 187 |
| nr | 28 639 | 9 428 | 14 621 |
| In all database | 28 992 | 9 544 | 14 648 |
Table 5 Single-gene function annotation statistics
| 数据库 Database | 注释的单基因数Number of annotated unigenes | 300 bp≤长度<1 000 bp 300 bp≤Length<1 000 bp | 长度≥1 000 bp Length≥1 000 bp |
|---|---|---|---|
| COG | 8 056 | 1 670 | 5 646 |
| GO | 17 733 | 5 300 | 9 680 |
| KEGG | 9 814 | 2 805 | 5 673 |
| KOG | 15 668 | 4 567 | 8 845 |
| Pfam | 18 933 | 5 043 | 12 068 |
| Swissprot | 19 579 | 5 863 | 11 158 |
| eggNOG | 26 275 | 8 298 | 14 187 |
| nr | 28 639 | 9 428 | 14 621 |
| In all database | 28 992 | 9 544 | 14 648 |
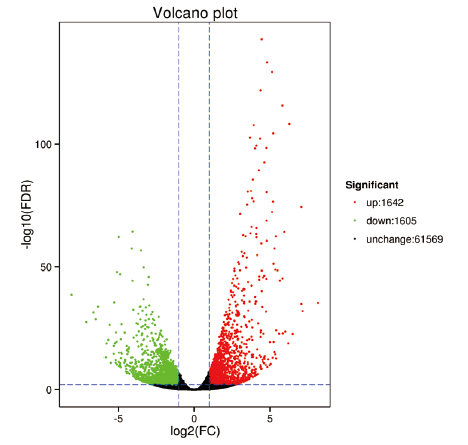
Fig. 3 Volcano map of differentially expressed genes The green and red dots in the graph indicate genes with significant expression differences,green indicates genes with down-regulated expression,red indicates genes with up-regulated expression,and black dots indicate genes without significant expression differences
| [1] | Plekhanova MN. Blue honeysuckle(Lonicera caerulea L.)- a new commercial berry crop for temperate climate:genetic resources and breeding[J]. Acta Hortic, 2000(538):159-164. |
| [2] | 霍俊伟, 杨国慧, 睢薇, 等. 蓝靛果忍冬(Lonicera caerulea)种质资源研究进展[J]. 园艺学报, 2005, 32(1):159-164. |
| Huo JW, Yang GH, Sui W, et al. Review of study on germplasm resources of blue honeysuckle(Lonicera caerulea L.)[J]. Acta Hortic Sin, 2005, 32(1):159-164. | |
| [3] |
Chaovanalikit A, Thompson MM, Wrolstad RE. Characterization and quantification of anthocyanins and polyphenolics in blue honeysuckle(Lonicera caerulea L.)[J]. J Agric Food Chem, 2004, 52(4):848-852.
doi: 10.1021/jf030509o URL |
| [4] |
Celli GB, Ghanem A, Brooks MSL. Optimization of ultrasound-assisted extraction of anthocyanins from haskap berries(Lonicera caerulea L.)using response surface methodology[J]. Ultrason Sonochem, 2015, 27:449-455.
doi: 10.1016/j.ultsonch.2015.06.014 URL |
| [5] | 刘奕琳. 蓝靛果花色苷分离及其抗氧化与抗癌功能研究[D]. 哈尔滨: 东北林业大学, 2012. |
| Liu YL. Sepaeration and reseach on antioxidant and anticancer anthocyanin of lonecera edulis[D]. Harbin: Northeast Forestry University, 2012. | |
| [6] |
Wang LS, Stoner GD. Anthocyanins and their role in cancer prevention[J]. Cancer Lett, 2008, 269(2):281-290.
doi: 10.1016/j.canlet.2008.05.020 URL |
| [7] | 张建全, 张含生, 张壮飞, 等. 小兴安岭地区蓝靛果优良品种引种栽培试验初报[J]. 农业科技通讯, 2018(1):152-155. |
| Zhang JQ, Zhang HS, Zhang ZF, et al. A preliminary report on the introduction and cultivation trials of good varieties of blue honeysuckle in the Xiaoxinganling region[J]. Bulletin of Agricultural Science and Technology, 2018(1):152-155. | |
| [8] |
Kong JM, Chia LS, Goh NK, et al. Analysis and biological activities of anthocyanins[J]. Phytochemistry, 2003, 64(5):923-933.
doi: 10.1016/S0031-9422(03)00438-2 URL |
| [9] |
Lohachoompol V, Mulholland M, Srzednicki G, et al. Determination of anthocyanins in various cultivars of highbush and rabbiteye blueberries[J]. Food Chem, 2008, 111(1):249-254.
doi: 10.1016/j.foodchem.2008.03.067 URL |
| [10] | 王忠华, 俞超, 吴月燕. 葡萄花青素苷生物合成研究进展[J]. 生物技术通报, 2012(9):1-7. |
| Wang ZH, Yu C, Wu YY. Progress on anthocyanin biosynthesis in grape[J]. Biotechnol Bull, 2012(9):1-7. | |
| [11] |
Springob K, Nakajima JI, Yamazaki M, et al. Recent advances in the biosynthesis and accumulation of anthocyanins[J]. Nat Prod Rep, 2003, 20(3):288-303.
pmid: 12828368 |
| [12] |
Zhou LP, Wang H, Yi JJ, et al. Anti-tumor properties of anthocyanins from Lonicera caerulea ‘Beilei’ fruit on human hepatocellular carcinoma:in vitro and in vivo study[J]. Biomed Pharmacother, 2018, 104:520-529.
doi: 10.1016/j.biopha.2018.05.057 URL |
| [13] | Pažereckaitė A, Jasutienė I, Šarkinas A, et al. Antimicrobial activity and composition of different cultivars of honeysuckle berry Lonicera caerulea L[C]// The 1st International Electronic Conference on Plant Science. Basel Switzerland: MDPI, 2020:1-15. |
| [14] |
Montefiori M, McGhie TK, Costa G, et al. Pigments in the fruit of red-fleshed kiwifruit(Actinidia chinensis and Actinidia deliciosa)[J]. J Agric Food Chem, 2005, 53(24):9526-9530.
doi: 10.1021/jf051629u URL |
| [15] |
Elitzur T, Yakir E, Quansah L, et al. Banana MaMADS transcription factors are necessary for fruit ripening and molecular tools to promote shelf-life and food security[J]. Plant Physiol, 2016, 171(1):380-391.
doi: 10.1104/pp.15.01866 pmid: 26956665 |
| [16] | 齐晓. 紫花苜蓿在转录组水平响应低温胁迫的差异表达基因研究[D]. 北京: 中国农业科学院, 2017. |
| Qi X. The analysis of the differentially expressed genes in alfalfa under cold stress at transcriptome level[D]. Beijing: Chinese Academy of Agricultural Sciences, 2017. | |
| [17] |
Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome[J]. Nat Biotechnol, 2011, 29(7):644-652.
doi: 10.1038/nbt.1883 pmid: 21572440 |
| [18] | 押辉远, 陈叶, 张岩松, 等. 槟榔不同发育时期果实转录组特征分析[J]. 热带作物学报, 2020, 41(7):1279-1287. |
| Ya HY, Chen Y, Zhang YS, et al. Analysis of transcriptome characteristics of Areca at different developmental stages[J]. Chin J Trop Crops, 2020, 41(7):1279-1287. | |
| [19] |
Raudsepp P, Anton D, Roasto M, et al. The antioxidative and antimicrobial properties of the blue honeysuckle(Lonicera caerulea L.), Siberian rhubarb(Rheum rhaponticum L.)and some other plants, compared to ascorbic acid and sodium nitrite[J]. Food Control, 2013, 31(1):129-135.
doi: 10.1016/j.foodcont.2012.10.007 URL |
| [20] |
Wu SS, He X, Wu XS, et al. Inhibitory effects of blue honeysuckle(Lonicera caerulea L)on adjuvant-induced arthritis in rats:Crosstalk of anti-inflammatory and antioxidant effects[J]. J Funct Foods, 2015, 17:514-523.
doi: 10.1016/j.jff.2015.06.007 URL |
| [21] | Zhao L, Li SR, Zhao L, et al. Antioxidant activities and major bioactive components of consecutive extracts from blue honeysuckle(Lonicera caerulea L.)cultivated in China[J]. J Food Biochem, 2015, 39(6):653-662. |
| [22] | 呼延文静. 蓝果忍冬种子及其籽油主要成分测定及抗氧化活性研究[D]. 哈尔滨: 东北农业大学, 2021. |
| Huyan WJ. Determination of main components and antioxidant activity of blue honeysuckle seeds and its seed oil[D]. Harbin: Northeast Agricultural University, 2021. | |
| [23] |
Huo JW, Liu P, Wang Y, et al. De novo transcriptome sequencing of blue honeysuckle fruit(Lonicera caerulea L.)and analysis of major genes involved in anthocyanin biosynthesis[J]. Acta Physiol Plant, 2016, 38(7):1-12.
doi: 10.1007/s11738-015-2023-4 URL |
| [24] | Yang Z, Chen H, Jia J, et al. De novo assembly and discovery of metabolic pathways and genes that are involved in defense against pests in Songyun Pinus massoniana Lamb[J]. Bangladesh J Bot, 2016, 45(4):855-863. |
| [25] |
Song HW, Zhao XX, Hu WC, et al. Comparative transcriptional analysis of loquat fruit identifies major signal networks involved in fruit development and ripening process[J]. Int J Mol Sci, 2016, 17(11):1837.
doi: 10.3390/ijms17111837 URL |
| [26] |
Li CQ, Wang Y, Huang XM, et al. De novo assembly and characterization of fruit transcriptome in Litchi chinensis Sonn and analysis of differentially regulated genes in fruit in response to shading[J]. BMC Genomics, 2013, 14:552.
doi: 10.1186/1471-2164-14-552 pmid: 23941440 |
| [27] | 徐伟君, 张九东, 郑博文, 等. 基于高通量测序的大叶女贞(Ligustrum lucidum)转录组分析[J]. 分子植物育种, 2019, 17(19):6295-6299. |
| Xu WJ, Zhang JD, Zheng BW, et al. Transcriptome analysis of Ligustrum lucidum based on high-throughput sequencing[J]. Mol Plant Breed, 2019, 17(19):6295-6299. | |
| [28] |
Li XY, Sun HY, Pei JB, et al. De novo sequencing and comparative analysis of the blueberry transcriptome to discover putative genes related to antioxidants[J]. Gene, 2012, 511(1):54-61.
doi: 10.1016/j.gene.2012.09.021 pmid: 22995346 |
| [29] |
赵阳阳, 郭雨潇, 张凌云. 文冠果果实转录组测序及分析[J]. 生物技术通报, 2019, 35(6):24-31.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-1101 URL |
| Zhao YY, Guo YX, Zhang LY. Transcriptome sequencing and analysis of Xanthoceras sorbifolia bunge fruit[J]. Biotechnol Bull, 2019, 35(6):24-31. | |
| [30] | 彭佳佳. 红掌转录组测序及花青素生物合成途径差异表达基因的分析[D]. 银川: 宁夏大学, 2015. |
| Peng JJ. Transcriptome analysis and anthocyanin biosynthesis genetic expression in the spathe of Anthurium andreanum[D]. Yinchuan: Ningxia University, 2015. | |
| [31] |
Dorcey E, Urbez C, Blázquez MA, et al. Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis[J]. Plant J, 2009, 58(2):318-332.
doi: 10.1111/j.1365-313X.2008.03781.x URL |
| [32] |
Groot SPC, Bruinsma J, Karssen CM. The role of endogenous gibberellin in seed and fruit development of tomato:studies with a gibberellin-deficient mutant[J]. Physiol Plant, 1987, 71(2):184-190.
doi: 10.1111/j.1399-3054.1987.tb02865.x URL |
| [33] | Stopar M. KNOCHE, Moritz, KHANAL, Bischnu P., STOPAR Matej. Russeting and microcracking of ‘Golden Delicious’ apple fruit concomitantly decline due to gibberellin A4+7 application[J]. J Am Soc Hortic Sci, 2011, 136(3):159-164. |
| [34] |
Serrani JC, Fos M, Atarés A, et al. Effect of gibberellin and auxin on parthenocarpic fruit growth induction in the cv micro-tom of tomato[J]. J Plant Growth Regul, 2007, 26(3):211-221.
doi: 10.1007/s00344-007-9014-7 URL |
| [35] | 赵荣华, 白世践, 陈光, 等. 赤霉素对无核白葡萄果实品质的影响[J]. 黑龙江农业科学, 2014(9):40-42. |
| Zhao RH, Bai SJ, Chen G, et al. Effect of gibberellin on fruit quality of thompsons seedless[J]. Heilongjiang Agric Sci, 2014(9):40-42. | |
| [36] |
Tiwari A, Offringa R, Heuvelink E. Auxin-induced fruit set in Capsicum annuum L. requires downstream gibberellin biosynthesis[J]. J Plant Growth Regul, 2012, 31(4):570-578.
doi: 10.1007/s00344-012-9267-7 URL |
| [37] |
Medina-Puche L, Cumplido-Laso G, Amil-Ruiz F, et al. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria x ananassa fruits[J]. J Exp Bot, 2014, 65(2):401-417.
doi: 10.1093/jxb/ert377 pmid: 24277278 |
| [38] |
Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits[J]. Trends Plant Sci, 2013, 18(9):477-483.
doi: 10.1016/j.tplants.2013.06.003 pmid: 23870661 |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [3] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [4] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [5] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [6] | ZHENG Huan, LIN Dong-mei, LIU Jun-yuan, ZHANG Yin-lian, LIN Biao-sheng, LIN Zhan-xi, LI Jing. Analysis of Amino Acid Metabolism Difference Between Fruiting Body and Mycelium of Taiwanofungus camphoratus by LC-QTOF-MS Metabonomics [J]. Biotechnology Bulletin, 2023, 39(5): 254-266. |
| [7] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [8] | ZHANG Le-le, WANG Guan, LIU Feng, HU Han-qiao, REN Lei. Isolation, Identification and Biocontrol Mechanism of an Antagonistic Bacterium Against Anthracnose on Mango Caused by Colletotrichum gloeosporioides [J]. Biotechnology Bulletin, 2023, 39(4): 277-287. |
| [9] | XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit [J]. Biotechnology Bulletin, 2023, 39(3): 152-162. |
| [10] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [11] | HU Li-li, LIN Bo-rong, WANG Hong-hong, CHEN Jian-song, LIAO Jin-ling, ZHUO Kan. Transcriptome and Candidate Effectors Analysis of Pratylenchus brachyurus [J]. Biotechnology Bulletin, 2023, 39(3): 254-266. |
| [12] | SUN Yan-qiu, XIE Cai-yun, TANG Yue-qin. Construction and Mechanism Analysis of High-temperature Resistant Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(11): 226-237. |
| [13] | MAO Ke-xin, WANG Hai-rong, AN Miao, LIU Teng-fei, WANG Shi-jin, LI Jian, LI Guo-tian. Identification of GRAS Gene Family and Expression Analysis Under Low Temperature Stress in Actinidia chinensis [J]. Biotechnology Bulletin, 2023, 39(11): 297-307. |
| [14] | LIU Yuan-yuan, WEI Chuan-zheng, XIE Yong-bo, TONG Zong-jun, HAN Xing, GAN Bing-cheng, XIE Bao-gui, YAN Jun-jie. Characteristics of Class II Peroxidase Gene Expression During Fruiting Body Development and Stress Response in Flammulina filiformis [J]. Biotechnology Bulletin, 2023, 39(11): 340-349. |
| [15] | XU Jun, YE Yu-qing, NIU Ya-jing, HUANG He, ZHANG Meng-meng. Transcriptome Analysis of Rhizome Development in Chrysanthemum× × morifolium [J]. Biotechnology Bulletin, 2023, 39(10): 231-245. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||