








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (4): 114-123.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0572
Previous Articles Next Articles
AI Lu( ), CHEN Wen-hui, SHI Jing-hui, REN Zhi-yuan, SHEN Wen-qi, YANG Jia-ning, LUO Jian-mei(
), CHEN Wen-hui, SHI Jing-hui, REN Zhi-yuan, SHEN Wen-qi, YANG Jia-ning, LUO Jian-mei( ), WANG Min
), WANG Min
Received:2022-05-09
Online:2023-04-26
Published:2023-05-16
AI Lu, CHEN Wen-hui, SHI Jing-hui, REN Zhi-yuan, SHEN Wen-qi, YANG Jia-ning, LUO Jian-mei, WANG Min. Cloning and Expression of 11α Hydroxylase from Aspergillus ochraceus and Analysis of Key Amino Acid Sites[J]. Biotechnology Bulletin, 2023, 39(4): 114-123.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 酶切位点 Restriction site | 引物用途 Primer application |
|---|---|---|---|
| 11αAO-F | ATGCCCTTCTTCACTGGGCTTCTGGCGAT | 无 | 从赭曲霉cDNA扩增11α羟化酶基因 |
| 11αAO-R | CTACACAGTTAAACTCGCCATATCGATCTCCGCC | 无 | Amplification of 11α hydroxylase gene from cDNA of Aspergillus ochraceus |
| CYP68J5-F | GCCGCCAGTGTGCTGGAATTCATGCCATTTTTTA- CTGGTTTGTTG | EcoR I | 构建酿酒酵母表达CYP68J5重组菌株 Construction of the recombinant S. cerevisiae expressing CYP68J5 |
| CYP68J5-R | TACATGATGCGGCCCTCTAGATTAAACAGTCAA- AGAAGCCATATCAA | Xba I | |
| CYP68J5-F | CGGGATCCCATGAACATCAAGAAGTTCGCCAAG | BamH I | 构建毕赤酵母表达CYP68J5重组菌株 Construction of the recombinant P. pastorisexpressing CYP68J5 |
| CYP68J5-R | CGGAATTCTCACTTGTTGACGGTGAGCTGCC | EcoR I | |
| CYP68J5-F | CGGAATTCATGCCGTTCTTCACCGGCC | EcoR I | 构建紫红红球菌表达CYP68J5重组菌株 |
| CYP68J5-R | GCTCTAGATCTAGATTAGACGGTGAGCGAGGCC | Xba I | Construction of the recombinant R. rhodochrous expressing CYP68J5 |
| CYP68J5_W89A-F | GCGGGTTCTTTGATTGTTTTGCCAC | 无 | 构建CYP68J5突变体 |
| CYP68J5_W89A-R | TTCAGACATCAATCTATATGGTTCATGAC | 无 | Construction of mutants of CYP68J5 |
| CYP68J5_F111A-F | GCTGAAACTCCAACTACTGATGATTCTC | 无 | |
| CYP68J5_F111 A-R | ATCCATTCTTGGATCATTTCTCAATTC | 无 | |
| CYP68J5_T115A-F | GCTACTGATGATTCTCATGGTTATATTCC | 无 | |
| CYP68J5_T115A-R | TGGAGTTTCAAAATCCATTCTTGGATC | 无 | |
| CYP68J5_D118A-F | GCTTCTCATGGTTATATTCCTGGTTTTG | 无 | |
| CYP68J5_D118A-R | ATCAGTAGTTGGAGTTTCAAAATCCAT | 无 | |
| CYP68J5_S119A-F | GCTCATGGTTATATTCCTGGTTTTG | 无 | |
| CYP68J5_S119A-R | ATCATCAGTAGTTGGAGTTTCAAAATC | 无 | |
| CYP68J5_L129A-F | GCAAATGCTGATCCAAATTTGACTAAAG | 无 | |
| CYP68J5_L129A-R | AGCATCAAAACCAGGAATATAACCATG | 无 | |
| CYP68J5_F216A-F | GCTGGTGTTGGTGATAAATTGAGAAT | 无 | |
| CYP68J5_F216A-R | AGCCAAAGCAGCATATTGAGAAGAAGTT | 无 | |
| CYP68J5_R223A-F | GCAATTTATCCAAGAATGATTAGACC | 无 | |
| CYP68J5_R223A-R | CAATTTATCACCAACACCAAAAGCC | 无 | |
| CYP68J5_I303A-F | GCTGTTGCTATTCATACTACTTCTG | 无 | |
| CYP68J5_I303A-R | AGACAAAGTAACTTGTTTCAAAACAG | 无 | |
| CYP68J5_V304A-F | GCTGCTATTCATACTACTTCTGATTTGT | 无 | |
| CYP68J5_V304A-R | AATAGACAAAGTAACTTGTTTCAAAAC | 无 | |
| CYP68J5_T308A-F | GCTACTTCTGATTTGTTATTGCAAGCT | 无 | |
| CYP68J5_T308A-R | ATGAATAGCAACAATAGACAAAGTAAC | 无 | |
| CYP68J5_L368A-F | GCGTTGGGATCTTTCAGAAGACAAG | 无 | |
| CYP68J5_L368A-R | AGTTGGTCTCAATCTTTGAGATTCTTTC | 无 | |
| CYP68J5_R373A-F | GCAAGACAAGCTACTAATGATATTAAATT | 无 | |
| CYP68J5_R373A-R | GAAAGATCCCAACAAAGTTGGTCTC | 无 | |
| CYP68J5_M488A-F | GCGACTTATTTGGCTGATCCAAACAC | 无 | |
| CYP68J5_M488A-R | ACCAATATTCAATGGTTGTGGTTTAAAAC | 无 | |
| CYP68J5_T489A-F | GCTTATTTGGCTGATCCAAACACTAG | 无 | |
| CYP68J5_T489A-R | CATACCAATATTCAATGGTTGTGGTTT | 无 |
Table 1 Primers used in this study
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 酶切位点 Restriction site | 引物用途 Primer application |
|---|---|---|---|
| 11αAO-F | ATGCCCTTCTTCACTGGGCTTCTGGCGAT | 无 | 从赭曲霉cDNA扩增11α羟化酶基因 |
| 11αAO-R | CTACACAGTTAAACTCGCCATATCGATCTCCGCC | 无 | Amplification of 11α hydroxylase gene from cDNA of Aspergillus ochraceus |
| CYP68J5-F | GCCGCCAGTGTGCTGGAATTCATGCCATTTTTTA- CTGGTTTGTTG | EcoR I | 构建酿酒酵母表达CYP68J5重组菌株 Construction of the recombinant S. cerevisiae expressing CYP68J5 |
| CYP68J5-R | TACATGATGCGGCCCTCTAGATTAAACAGTCAA- AGAAGCCATATCAA | Xba I | |
| CYP68J5-F | CGGGATCCCATGAACATCAAGAAGTTCGCCAAG | BamH I | 构建毕赤酵母表达CYP68J5重组菌株 Construction of the recombinant P. pastorisexpressing CYP68J5 |
| CYP68J5-R | CGGAATTCTCACTTGTTGACGGTGAGCTGCC | EcoR I | |
| CYP68J5-F | CGGAATTCATGCCGTTCTTCACCGGCC | EcoR I | 构建紫红红球菌表达CYP68J5重组菌株 |
| CYP68J5-R | GCTCTAGATCTAGATTAGACGGTGAGCGAGGCC | Xba I | Construction of the recombinant R. rhodochrous expressing CYP68J5 |
| CYP68J5_W89A-F | GCGGGTTCTTTGATTGTTTTGCCAC | 无 | 构建CYP68J5突变体 |
| CYP68J5_W89A-R | TTCAGACATCAATCTATATGGTTCATGAC | 无 | Construction of mutants of CYP68J5 |
| CYP68J5_F111A-F | GCTGAAACTCCAACTACTGATGATTCTC | 无 | |
| CYP68J5_F111 A-R | ATCCATTCTTGGATCATTTCTCAATTC | 无 | |
| CYP68J5_T115A-F | GCTACTGATGATTCTCATGGTTATATTCC | 无 | |
| CYP68J5_T115A-R | TGGAGTTTCAAAATCCATTCTTGGATC | 无 | |
| CYP68J5_D118A-F | GCTTCTCATGGTTATATTCCTGGTTTTG | 无 | |
| CYP68J5_D118A-R | ATCAGTAGTTGGAGTTTCAAAATCCAT | 无 | |
| CYP68J5_S119A-F | GCTCATGGTTATATTCCTGGTTTTG | 无 | |
| CYP68J5_S119A-R | ATCATCAGTAGTTGGAGTTTCAAAATC | 无 | |
| CYP68J5_L129A-F | GCAAATGCTGATCCAAATTTGACTAAAG | 无 | |
| CYP68J5_L129A-R | AGCATCAAAACCAGGAATATAACCATG | 无 | |
| CYP68J5_F216A-F | GCTGGTGTTGGTGATAAATTGAGAAT | 无 | |
| CYP68J5_F216A-R | AGCCAAAGCAGCATATTGAGAAGAAGTT | 无 | |
| CYP68J5_R223A-F | GCAATTTATCCAAGAATGATTAGACC | 无 | |
| CYP68J5_R223A-R | CAATTTATCACCAACACCAAAAGCC | 无 | |
| CYP68J5_I303A-F | GCTGTTGCTATTCATACTACTTCTG | 无 | |
| CYP68J5_I303A-R | AGACAAAGTAACTTGTTTCAAAACAG | 无 | |
| CYP68J5_V304A-F | GCTGCTATTCATACTACTTCTGATTTGT | 无 | |
| CYP68J5_V304A-R | AATAGACAAAGTAACTTGTTTCAAAAC | 无 | |
| CYP68J5_T308A-F | GCTACTTCTGATTTGTTATTGCAAGCT | 无 | |
| CYP68J5_T308A-R | ATGAATAGCAACAATAGACAAAGTAAC | 无 | |
| CYP68J5_L368A-F | GCGTTGGGATCTTTCAGAAGACAAG | 无 | |
| CYP68J5_L368A-R | AGTTGGTCTCAATCTTTGAGATTCTTTC | 无 | |
| CYP68J5_R373A-F | GCAAGACAAGCTACTAATGATATTAAATT | 无 | |
| CYP68J5_R373A-R | GAAAGATCCCAACAAAGTTGGTCTC | 无 | |
| CYP68J5_M488A-F | GCGACTTATTTGGCTGATCCAAACAC | 无 | |
| CYP68J5_M488A-R | ACCAATATTCAATGGTTGTGGTTTAAAAC | 无 | |
| CYP68J5_T489A-F | GCTTATTTGGCTGATCCAAACACTAG | 无 | |
| CYP68J5_T489A-R | CATACCAATATTCAATGGTTGTGGTTT | 无 |
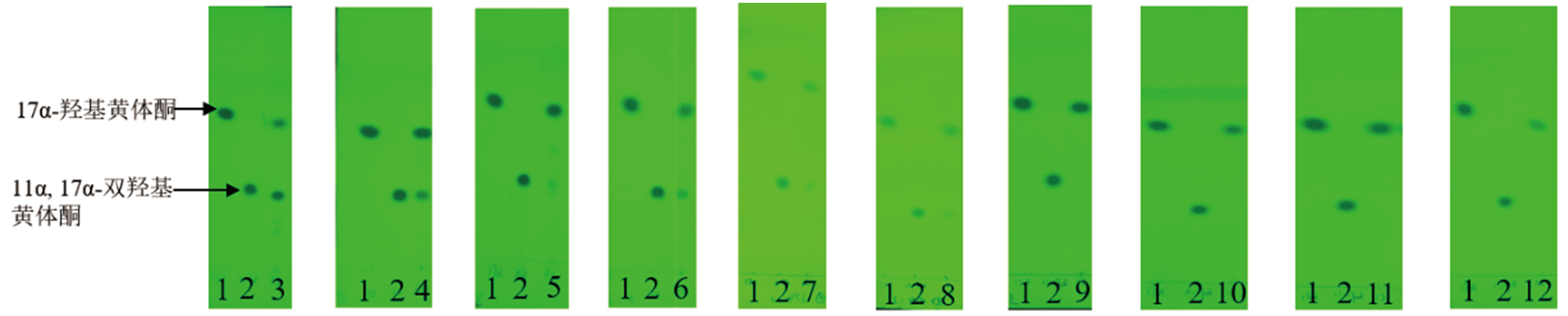
Fig. 2 TLC results of 17α-hydroxyprogesterone at 48 h by 10 molds 1:Standard of 17α-hydroxyprogesterone. 2:Standard of 11α,17α-dihydroxy progesterone. 3:Aspergillus ochraceus. 4:Cunningpamycetes elegans. 5:Absidia coerulea. 6:Beauveria bassiana. 7:Metarhizium anisopliae. 8:Aspergillus niger. 9:Penicillium citreoviridin. 10:Aspergillus flavus. 11:Penicillium citrinum. 12:Penicillium raistrickii
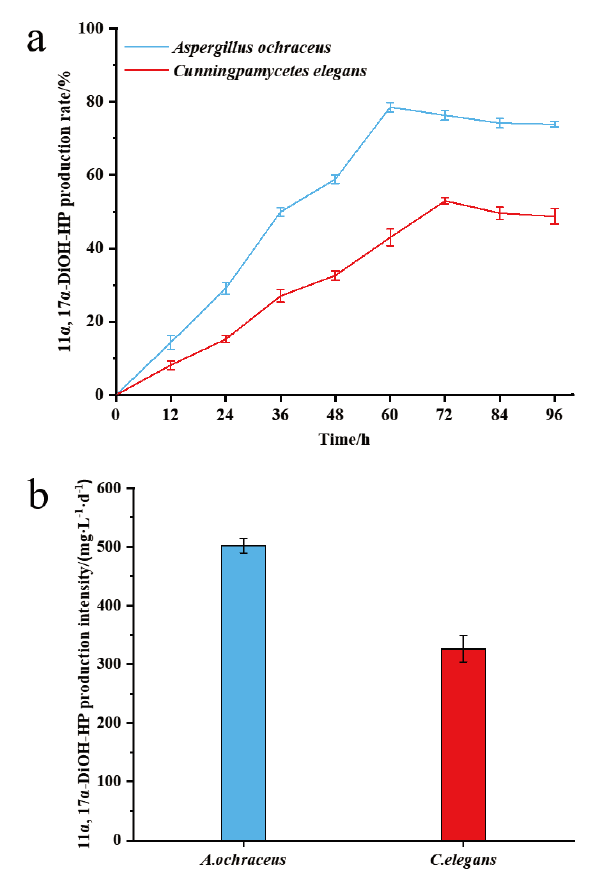
Fig. 3 Molar production rate(a)and production intensity(b)of 11α,17α-dihydroxy progesterone from 17α-hydroxyprogesterone by Aspergillus ochraceus and Cunningpamycetes elegans,respectively

Fig. 4 Verification of the recombinant P. pastoris(a),S. cerevisiae(b)and R. rhodochrous(c)expressing CYP68J5 M:DL 2000 DNA marker;1:PCR product of pYES2-11αAo;2:PCR product of pNV18-11αAo
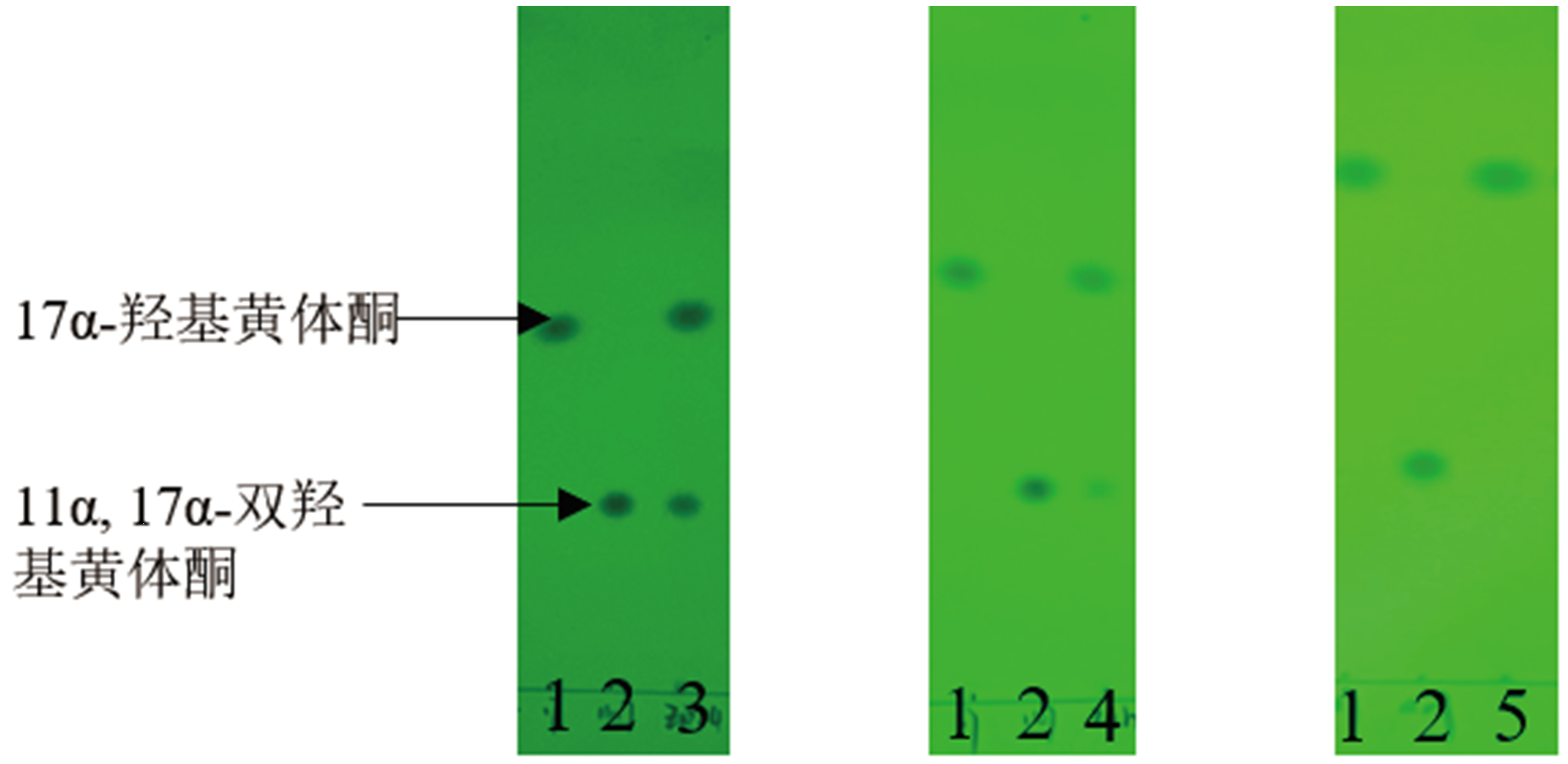
Fig. 5 TLC biotransformation results of 17α-hydroxypro-gesterone at 48 h by the recombinant S. cerevisiae,P. pastoris and R. rhodochrous expressing CYP68J5,respectively 1:Standard of 17α-hydroxyprogesterone. 2. Standard of 11α,17α-dihydroxy progesterone. 3:Saccharomyces cerevisiae. 4:Pichia pastoris. 5:Rhodococcus rhodochrous
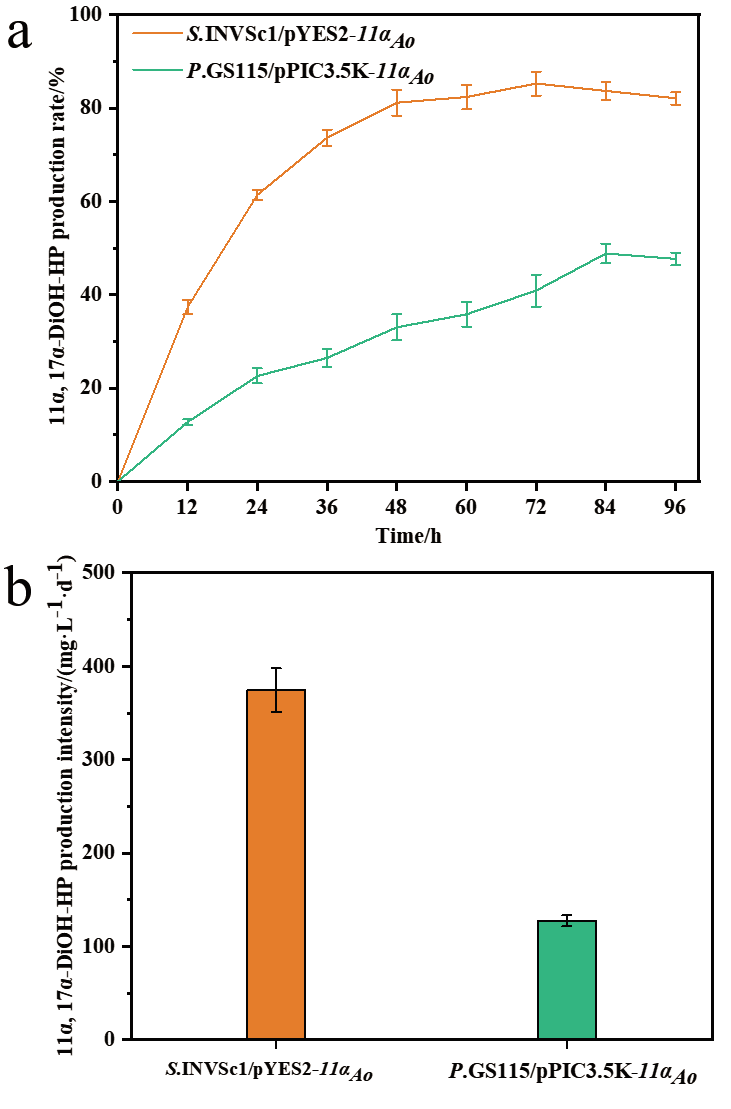
Fig. 6 Molar production rate(a)and production inten-sity(b)of 11α, 17α-dihydroxy progesterone from 17α-hydroxyprogesterone by the recombinant S. cer-evisiae and P. pastoris expressing CYP68J5, respec-tively

Fig. 8 Predicted overall structure of CYP68J5 and the molecular docking with 17α-hydroxyprogesterone α helix,β folding and random curl are colored in bright blue,bright purple and pink,respectively. The active site residues and the substrate are colored in bright yellow and dark red,respectively
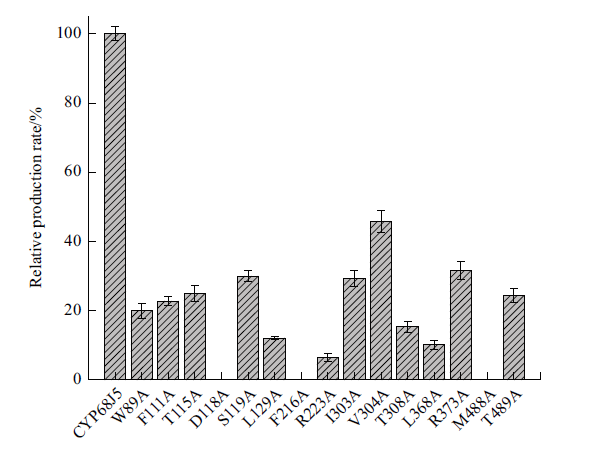
Fig. 9 Relative molar production rate of 11α, 17α-dihydr-oxy progesterone from 17α-hydroxyprogesterone by the mutants compared to that of the wild-type CYP68J5 The data represent the mean value of three independent experiments. The relative molar production rate is calculated by using the molar production rate of wild-type CYP68J5 as 100%

Fig. 10 Local model of the key amino acid site residues D118(a),F216(b)and M488(c)of CYP68J5 and the alanine mutation site residues A118(d),A216(e)and A488(f)
| [1] |
Meister PD, Peterson DH, Murray HC, et al. Microbiological transformations of steroids V the oxygenation of 17α-hydroxyprogesterone to 6β, 17α-dihydroxyprogesterone and 11α, 17α-Dihydroxyprogesterone1[J]. J Am Chem Soc, 1953, 75(2):416-418.
doi: 10.1021/ja01098a046 URL |
| [2] | Alejandro Z, Rubin BA. Process for the introduction of an 11 alpha-oh group into cyclopentanophenanthrene derivatives by cunninghamella echinulata: US2812286[P]. 1957-11-05. |
| [3] | Žakelj-Mavrič M, Belič I. Hydroxylation of steroids with 11α-hydroxylase of Rhizopus nigricans[J]. J Steroid Biochem, 1987, 28(2):197-201. |
| [4] | 王建, 等. 11α, 17α-二羟基-雄甾-4-烯-3, 20-二酮的制备方法:CN102061320A[P]. 2011-05-18. |
| Wang J, et al. Preparation method of 11 alpha, 17 alpha-dyhydroxyl-androst-4-ene-3, 20-Dione:CN102061320A[P]. 2011-05-18 | |
| [5] |
Koehn EM, Kohen A. Flavin-dependent thymidylate synthase:a novel pathway towards thymine[J]. Arch Biochem Biophys, 2010, 493(1):96-102.
doi: 10.1016/j.abb.2009.07.016 pmid: 19643076 |
| [6] | Hannemann F, Bichet A, Ewen KM, et al. Cytochrome P450 systems—biological variations of electron transport chains[J]. Biochim Biophys Acta BBA Gen Subj, 2007, 1770(3):330-344. |
| [7] | Črešnar B, Petrič Š. Cytochrome P450 enzymes in the fungal kingdom[J]. Biochim Biophys Acta BBA Proteins Proteom, 2011, 1814(1):29-35. |
| [8] |
Park J, Lee S, Choi J, et al. Fungal cytochrome P450 database[J]. BMC Genomics, 2008, 9:402.
doi: 10.1186/1471-2164-9-402 pmid: 18755027 |
| [9] | 苏珊尼·博尔藤, 罗伯特·克莱顿, 阿伦·伊斯顿, 等. 赭曲霉11α-羟化酶和氧化还原酶:CN1531590A[P]. 2004-09-22. |
| Bolten S, Clayton R, Easton A, et al. Aspergillus ocharceus 11 alpha hydroxylase and oxidoreductase:CN1531590A[P]. 2004-09-22 | |
| [10] |
Wang X, Yang XW, Jia X, et al. Determination of steroid hydroxylation specificity of an industrial strain Aspergillus ochraceus TCCC41060 by cytochrome P450 gene CYP68J5[J]. Ann Microbiol, 2020, 70:45.
doi: 10.1186/s13213-020-01577-6 |
| [11] |
Wang RJ, Sui PC, Hou XJ, et al. Cloning and identification of a novel steroid 11α-hydroxylase gene from Absidia coerulea[J]. J Steroid Biochem Mol Biol, 2017, 171:254-261.
doi: 10.1016/j.jsbmb.2017.04.006 URL |
| [12] |
Petrič S, Hakki T, Bernhardt R, et al. Discovery of a steroid 11α-hydroxylase from Rhizopus oryzae and its biotechnological application[J]. J Biotechnol, 2010, 150(3):428-437.
doi: 10.1016/j.jbiotec.2010.09.928 pmid: 20850485 |
| [13] | 林本凤, 职亚飞, 刘晓光, 等. 黑曲霉ATCC1015催化16α, 17α-环氧黄体酮11α-羟基化及相关P450基因诱导表达[J]. 天津科技大学学报, 2017, 32(6):8-14. |
| Lin BF, Zhi YF, Liu XG, et al. 11α-hydroxylation of 16α, 17α-epoxy progesterone by Aspergillus niger ATCC1015 and induction expression of relevant cytochromes P450 genes[J]. J Tianjin Univ Sci Technol, 2017, 32(6):8-14. | |
| [14] |
Jiao S, Yu HM, Shen ZY. Core element characterization of Rho-dococcus promoters and development of a promoter-RBS mini-pool with different activity levels for efficient gene expression[J]. N Biotechnol, 2018, 44:41-49.
doi: 10.1016/j.nbt.2018.04.005 URL |
| [15] | 乔玉茜. 17α羟基黄体酮11α羟化菌株筛选及其转化工艺研究[D]. 天津: 天津科技大学, 2017. |
| Qiao YQ. Screeing and transformation conditions of 11α-hydroxylation of 17α-hydroxy progesterone[D]. Tianjin: Tianjin University of Science & Technology, 2017 | |
| [16] | 牟晓然. 赭曲霉制备心血管药物C11α羟基化坎利酮培养条件的研究[D]. 长春: 吉林大学, 2008. |
| Mou XR. Study on the cultural condition of the cardiovascular drugs canrenone into C11α-hydroxylated product by Aspergillus ochraceus[D]. Changchun: Jilin University, 2008 | |
| [17] | 龙尾, 等. 赭曲霉对16α, 17α-环氧黄体酮的C11α-羟基化工艺研究[J]. 化学与生物工程, 2010, 27(8):44-47. |
| Long W, et al. Study on C11α-hydroxylation of 16α, 17α-epoxyprogesterone by Aspergillus ochraceus 405[J]. Chem Bioeng, 2010, 27(8):44-47. | |
| [18] | 徐银, 等. 雅致小克银汉霉对16α, 17α-环氧黄体酮C11α-羟基化的工艺研究[J]. 中国生化药物杂志, 2009, 30(4):239-242. |
| Xu Y, et al. Studies on the fermentation conditions of C11α-hydroxylation of 16α, 17α-epoxy-4-pregnene-3, 20-Dione by Cun-ninghamella elegans[J]. Chin J Biochem Pharm, 2009, 30(4):239-242. | |
| [19] | 刘玲玲, 等. 金龟子绿僵菌对甾体底物11-α羟化反应的工艺[J]. 食品与生物技术学报, 2006, 25(5):77-80, 87. |
| Liu LL, et al. 11-αhydroxylation process of steroids with Metarhi-zium anisopliae[J]. J Food Sci Biotechnol, 2006, 25(5):77-80, 87. | |
| [20] | 魏琦. 雄烯二酮11α羟化的球孢白僵菌菌株筛选及其转化工艺研究[D]. 武汉: 华中农业大学, 2007. |
| Wei Q. Screeing and transformation conditions of 11α-hydroxylation of androst-4-ene-3, 17-Dione by Beauveria bassiana[D]. Wuhan: Huazhong Agricultural University, 2007 | |
| [21] | 申雁冰, 王敏, 骆健美, 等. 一种利用微生物转化生产7α-羟基-脱氢表雄酮的方法:CN110885869A[P]. 2020-03-17. |
| Shen YB, Wang M, Luo JM, et al. Method for conversion production of 7alpha-OH-DHEA by using microorganism:CN110885869A[P]. 2020-03-17 | |
| [22] | 王敏, 申雁冰, 薛玮莹, 等. 一株蓝色犁头霉及其应用:CN107475131B[P]. 2021-01-15. |
| Wang M, Shen YB, Xu WY, et al. Absidia coerulea and application thereof:CN107475131B[P]. 2021-01-15. | |
| [23] | 李雪龙, 等. 增加雷斯青霉15α-羟化酶基因拷贝数提高甾体转化效率[J]. 天津科技大学学报, 2019, 34(4):16-21, 29. |
| Li XL, et al. Increasing 15α-hydroxylase gene copy number in Peni-cillium raistrickii to enhance the transformation efficiency of ster-oid[J]. J Tianjin Univ Sci Technol, 2019, 34(4):16-21, 29. | |
| [24] |
Chen J, Fan FY, Qu G, et al. Identification of Absidia orchidis steroid 11β-hydroxylation system and its application in engineering Saccharomyces cerevisiae for one-step biotransformation to produce hydrocortisone[J]. Metab Eng, 2020, 57:31-42.
doi: 10.1016/j.ymben.2019.10.006 URL |
| [25] |
Chen J, Tang JL, Xi YY, et al. Production of 14α-hydroxysteroids by a recombinant Saccharomyces cerevisiae biocatalyst expressing of a fungal steroid 14α-hydroxylation system[J]. Appl Microbiol Biotechnol, 2019, 103(20):8363-8374.
doi: 10.1007/s00253-019-10076-x |
| [26] |
Jia LG, Dong JZ, Wang RJ, et al. Identification and characterization of the steroid 15α-hydroxylase gene from Penicillium raistrickii[J]. Appl Microbiol Biotechnol, 2017, 101(16):6409-6418.
doi: 10.1007/s00253-017-8377-3 URL |
| [27] | Lu W, et al. A fungal P450 enzyme from Thanatephorus cucumeris with steroid hydroxylation capabilities[J]. Appl Environ Microbiol, 2018, 84(13):e00503-e00518. |
| [28] | Lu W, Feng JH, Chen X, et al. Distinct regioselectivity of fungal P450 enzymes for steroidal hydroxylation[J]. Appl Environ Microbiol, 2019, 85(18):e01182-19. |
| [29] |
Yasutake Y, et al. A single mutation at the ferredoxin binding site of P450 Vdh enables efficient biocatalytic production of 25-hydroxyvitamin D3[J]. Chembiochem, 2013, 14(17):2284-2291.
doi: 10.1002/cbic.v14.17 URL |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||