








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (12): 1-10.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0014
LI Xiao-fan1,2( ), GENG Dan-dan1,2, BI Yu-lin2, JIANG Yong2, WANG Zhi-xiu2, CHANG Guo-bin1,2, CHEN Guo-hong1,2, BAI Hao1(
), GENG Dan-dan1,2, BI Yu-lin2, JIANG Yong2, WANG Zhi-xiu2, CHANG Guo-bin1,2, CHEN Guo-hong1,2, BAI Hao1( )
)
Received:2022-01-05
Online:2022-12-26
Published:2022-12-29
Contact:
BAI Hao
E-mail:ylcccoco@126.com;bhowen1027@yzu.edu.cn
LI Xiao-fan, GENG Dan-dan, BI Yu-lin, JIANG Yong, WANG Zhi-xiu, CHANG Guo-bin, CHEN Guo-hong, BAI Hao. Research Progress in Unconventional miRNA Functions[J]. Biotechnology Bulletin, 2022, 38(12): 1-10.
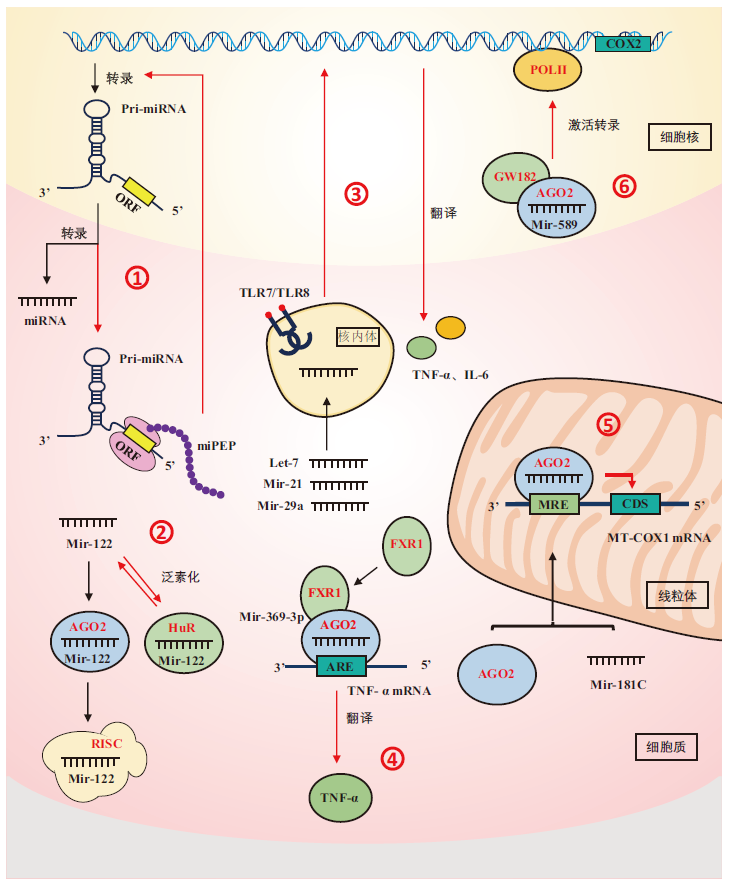
Fig. 1 Unconventional miRNA functions ①:pri-miRNAs coding for peptides miPEPs;②:miRNAs interacting with non-AGO proteins;③:miRNAs as signals molecules activating TLR receptors;④:miRNAs upregulating protein expression;⑤:miRNAs regulating the gene mRNA related to mitochondria in target;⑥:miRNAs directly regulating gene transcription
| 作用机制Functions | 物种Species | 性状Traits | 相关miRNAR Related miRNAs | 参考文献Reference |
|---|---|---|---|---|
| miRNA前体pri-miRNA编码调节肽miPEPs | 植物 | 根系发育 | pri-miR176、pri-miR165a | [ |
| pri-mir171d | [ | |||
| pri-mir858a | [ | |||
| 植物 | 叶片发育 | mir-165a | [ | |
| 植物 | 大豆根瘤结瘤 | mir-172c | [ | |
| 人类 | 前列腺癌 | miR-200a、miR-200b | [ | |
| 人类 | 类银屑病与多发性硬化症(EAE) | mir-155 | [ | |
| miRNA与多种功能蛋白相结合 | 人类 | 饥饿应激反应 | mir-122 | [ |
| 人类 | 慢性骨髓性白血病 | mir-328 | [ | |
| 人类 | 癌细胞发育调节 | mir-346、mir-138、pri-let-7 | [ | |
| [ | ||||
| 动物 | 哺乳动物胚胎发育 | let-7 | [ | |
| miRNA作为信号分子直接激活TLR受体蛋白 | 小鼠 | 阿尔茨海默病 | let-7 | [ |
| 小鼠 | 中枢神经系统疾病 | mir-20a-5p、mir-148b-3p | [ | |
| miRNA上调蛋白表达水平 | 人类 | 丙型肝炎病毒(HCV) | mir-122 | [ |
| 人类 | 巨噬细胞调节 | mir-125b | [ | |
| 人类 | 宫颈癌 | mir-346 | [ | |
| 人类 | 小儿癌症 | mir-483 | [ | |
| 动物 | 肠道菌群调节 | gga-miR-222a | [ | |
| 植物 | 干旱胁迫 | mir-156 | [ | |
| miRNA靶向调控线粒体相关基因mRNA | 人类 | 骨骼肌线粒体调节 | pre-mir-302a、pre-let-7b、mir-365 | [ |
| 大鼠 | 心脏线粒体调节 | mir-181c | [ | |
| 人类 | 肿瘤细胞代谢和化学耐药性 | mir-2392 | [ | |
| 小鼠 | 心肌线粒体调节 | mir-1 | [ | |
| 人类 | 神经发育 | mir-338 | [ | |
| F:miRNA直接调控基因转录过程 | 小鼠 | 肿瘤生长调节 | mir-744、mir-1186 | [ |
| 小鼠 | 细胞凋亡调节 | mir-709 | [ | |
| 人类 | 肿瘤生长调节 | miR-24、miR-16a-1 | [ |
Table 1 Research progresses in unconventional miRNA functions
| 作用机制Functions | 物种Species | 性状Traits | 相关miRNAR Related miRNAs | 参考文献Reference |
|---|---|---|---|---|
| miRNA前体pri-miRNA编码调节肽miPEPs | 植物 | 根系发育 | pri-miR176、pri-miR165a | [ |
| pri-mir171d | [ | |||
| pri-mir858a | [ | |||
| 植物 | 叶片发育 | mir-165a | [ | |
| 植物 | 大豆根瘤结瘤 | mir-172c | [ | |
| 人类 | 前列腺癌 | miR-200a、miR-200b | [ | |
| 人类 | 类银屑病与多发性硬化症(EAE) | mir-155 | [ | |
| miRNA与多种功能蛋白相结合 | 人类 | 饥饿应激反应 | mir-122 | [ |
| 人类 | 慢性骨髓性白血病 | mir-328 | [ | |
| 人类 | 癌细胞发育调节 | mir-346、mir-138、pri-let-7 | [ | |
| [ | ||||
| 动物 | 哺乳动物胚胎发育 | let-7 | [ | |
| miRNA作为信号分子直接激活TLR受体蛋白 | 小鼠 | 阿尔茨海默病 | let-7 | [ |
| 小鼠 | 中枢神经系统疾病 | mir-20a-5p、mir-148b-3p | [ | |
| miRNA上调蛋白表达水平 | 人类 | 丙型肝炎病毒(HCV) | mir-122 | [ |
| 人类 | 巨噬细胞调节 | mir-125b | [ | |
| 人类 | 宫颈癌 | mir-346 | [ | |
| 人类 | 小儿癌症 | mir-483 | [ | |
| 动物 | 肠道菌群调节 | gga-miR-222a | [ | |
| 植物 | 干旱胁迫 | mir-156 | [ | |
| miRNA靶向调控线粒体相关基因mRNA | 人类 | 骨骼肌线粒体调节 | pre-mir-302a、pre-let-7b、mir-365 | [ |
| 大鼠 | 心脏线粒体调节 | mir-181c | [ | |
| 人类 | 肿瘤细胞代谢和化学耐药性 | mir-2392 | [ | |
| 小鼠 | 心肌线粒体调节 | mir-1 | [ | |
| 人类 | 神经发育 | mir-338 | [ | |
| F:miRNA直接调控基因转录过程 | 小鼠 | 肿瘤生长调节 | mir-744、mir-1186 | [ |
| 小鼠 | 细胞凋亡调节 | mir-709 | [ | |
| 人类 | 肿瘤生长调节 | miR-24、miR-16a-1 | [ |
| [1] |
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene Lin-4 encodes small RNAs with antisense complementarity to Lin-14[J]. Cell, 1993, 75(5):843-854.
doi: 10.1016/0092-8674(93)90529-y pmid: 8252621 |
| [2] |
Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans[J]. Nature, 2000, 403(6772):901-906.
doi: 10.1038/35002607 URL |
| [3] |
Stavast C, Erkeland S. The non-canonical aspects of microRNAs:many roads to gene regulation[J]. Cells, 2019, 8(11):1465.
doi: 10.3390/cells8111465 URL |
| [4] |
Partin AC, Zhang KM, Jeong BC, et al. Cryo-EM structures of human drosha and DGCR8 in complex with primary microRNA[J]. Mol Cell, 2020, 78(3):411-422. e4.
doi: S1097-2765(20)30109-X pmid: 32220646 |
| [5] |
Rice GM, Shivashankar V, Ma EJ, et al. Functional atlas of primary miRNA maturation by the microprocessor[J]. Mol Cell, 2020, 80(5):892-902. e4.
doi: 10.1016/j.molcel.2020.10.028 pmid: 33188727 |
| [6] |
Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs[J]. RNA, 2004, 10(2):185-191.
pmid: 14730017 |
| [7] |
Jones-Rhoades MW, Bartel DP, Bartel B. Micrornas and their regulatory roles in plants[J]. Annu Rev Plant Biol, 2006, 57:19-53.
pmid: 16669754 |
| [8] |
Wong JJL, Ritchie W, Gao DD, et al. Identification of nuclear-enriched miRNAs during mouse granulopoiesis[J]. J Hematol Oncol, 2014, 7:42.
doi: 10.1186/1756-8722-7-42 URL |
| [9] |
Dragomir MP, Knutsen E, Calin GA. SnapShot:unconventional miRNA functions[J]. Cell, 2018, 174(4):1038-1038. e1.
doi: S0092-8674(18)30968-1 pmid: 30096304 |
| [10] |
Huffaker A, Dafoe NJ, Schmelz EA. ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance[J]. Plant Physiol, 2011, 155(3):1325-1338.
doi: 10.1104/pp.110.166710 pmid: 21205619 |
| [11] |
Couzigou JM, André O, Guillotin B, et al. Use of microRNA-encoded peptide miPEP172c to stimulate nodulation in soybean[J]. New Phytol, 2016, 211(2):379-381.
doi: 10.1111/nph.13991 URL |
| [12] |
Yamaguchi Y, Huffaker A, Bryan AC, et al. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis[J]. Plant Cell, 2010, 22(2):508-522.
doi: 10.1105/tpc.109.068874 URL |
| [13] |
Oh E, Seo PJ, Kim J. Signaling peptides and receptors coordinating plant root development[J]. Trends Plant Sci, 2018, 23(4):337-351.
doi: S1360-1385(17)30283-2 pmid: 29366684 |
| [14] |
Ormancey M le Ru A, Duboé C, et al. Internalization of miPEP165a into Arabidopsis roots depends on both passive diffusion and endocytosis-associated processes[J]. Int J Mol Sci, 2020, 21(7):2266.
doi: 10.3390/ijms21072266 URL |
| [15] |
Lauressergues D, Couzigou JM, Clemente HS, et al. Primary transcripts of microRNAs encode regulatory peptides[J]. Nature, 2015, 520(7545):90-93.
doi: 10.1038/nature14346 URL |
| [16] |
Chen QJ, Deng BH, Gao J, et al. A miRNA-encoded small peptide, vvi-miPEP171d1, regulates adventitious root formation[J]. Plant Physiol, 2020, 183(2):656-670.
doi: 10.1104/pp.20.00197 URL |
| [17] |
Wang JZ, Zhu S, Meng N, et al. ncRNA-encoded peptides or proteins and cancer[J]. Mol Ther, 2019, 27(10):1718-1725.
doi: S1525-0016(19)30402-2 pmid: 31526596 |
| [18] |
Fang JB, Morsalin S, Rao V, et al. Decoding of non-coding DNA and non-coding RNA:pri-micro RNA-encoded novel peptides regulate migration of cancer cells[J]. J Pharmaceut Sci Pharmacol, 2017, 3(1):23-27.
doi: 10.1166/jpsp.2017.1070 URL |
| [19] | Niu LM, Lou FZ, Sun Y, et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation[J]. Sci Adv, 2020, 6(21):eaaz2059. |
| [20] |
Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins[J]. Nat Rev Genet, 2014, 15(12):829-845.
doi: 10.1038/nrg3813 pmid: 25365966 |
| [21] |
Lunde BM, Moore C, Varani G. RNA-binding proteins:modular design for efficient function[J]. Nat Rev Mol Cell Biol, 2007, 8(6):479-490.
doi: 10.1038/nrm2178 URL |
| [22] |
Glisovic T, Bachorik JL, Yong J, et al. RNA-binding proteins and post-transcriptional gene regulation[J]. FEBS Lett, 2008, 582(14):1977-1986.
doi: 10.1016/j.febslet.2008.03.004 pmid: 18342629 |
| [23] |
Meister G. Argonaute proteins:functional insights and emerging roles[J]. Nat Rev Genet, 2013, 14(7):447-459.
doi: 10.1038/nrg3462 pmid: 23732335 |
| [24] |
Kwon SC, Nguyen TA, Choi YG, et al. Structure of human DROSHA[J]. Cell, 2016, 164(1/2):81-90.
doi: 10.1016/j.cell.2015.12.019 URL |
| [25] |
Yoon JH, De S, Srikantan S, et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity[J]. Nat Commun, 2014, 5:5248.
doi: 10.1038/ncomms6248 URL |
| [26] |
Mukherjee N, Corcoran DL, Nusbaum JD, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability[J]. Mol Cell, 2011, 43(3):327-339.
doi: 10.1016/j.molcel.2011.06.007 pmid: 21723170 |
| [27] |
Janas MM, Wang BB, Harris AS, et al. Alternative RISC assembly:binding and repression of microRNA-mRNA duplexes by human Ago proteins[J]. RNA, 2012, 18(11):2041-2055.
doi: 10.1261/rna.035675.112 pmid: 23019594 |
| [28] | Zealy RW, Wrenn SP, Davila S, et al. microRNA-binding proteins:specificity and function[J]. Wiley Interdiscip Rev RNA, 2017, 8(5): 2017 Sep; 8(5). |
| [29] |
Mukherjee K, Ghoshal B, Ghosh S, et al. Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response[J]. EMBO Rep, 2016, 17(8):1184-1203.
doi: 10.15252/embr.201541930 pmid: 27402548 |
| [30] |
Eiring AM, Harb JG, Neviani P, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts[J]. Cell, 2010, 140(5):652-665.
doi: 10.1016/j.cell.2010.01.007 pmid: 20211135 |
| [31] |
Yoon JH, Abdelmohsen K, Kim J, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination[J]. Nat Commun, 2013, 4:2939.
doi: 10.1038/ncomms3939 URL |
| [32] |
Peredo J, Villacé P, Ortín J, et al. Human Staufen1 associates to miRNAs involved in neuronal cell differentiation and is required for correct dendritic formation[J]. PLoS One, 2014, 9(11):e113704.
doi: 10.1371/journal.pone.0113704 URL |
| [33] |
Song G, Wang RJ, Guo JF, et al. miR-346 and miR-138 competitively regulate hTERT in GRSF1- and AGO2-dependent manners, respectively[J]. Sci Rep, 2015, 5:15793.
doi: 10.1038/srep15793 pmid: 26507454 |
| [34] |
Piskounova E, Viswanathan SR, Janas M, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28[J]. J Biol Chem, 2008, 283(31):21310-21314.
doi: 10.1074/jbc.C800108200 pmid: 18550544 |
| [35] | Lehmann SM, Krüger C, Park B, et al. An unconventional role for miRNA:let-7 activates Toll-like receptor 7 and causes neurodegeneration[J]. Nat Neurosci, 2012, 15(6):827-835. |
| [36] |
Wu NM, Morsey BM, Emanuel KM, et al. Sequence-specific extracellular microRNAs activate TLR7 and induce cytokine secretion and leukocyte migration[J]. Mol Cell Biochem, 2021, 476(11):4139-4151.
doi: 10.1007/s11010-021-04220-3 pmid: 34313894 |
| [37] |
Henke JI, Goergen D, Zheng JF, et al. microRNA-122 stimulates translation of hepatitis C virus RNA[J]. EMBO J, 2008, 27(24):3300-3310.
doi: 10.1038/emboj.2008.244 pmid: 19020517 |
| [38] |
Vasudevan S. Posttranscriptional upregulation by microRNAs[J]. Wiley Interdiscip Rev RNA, 2012, 3(3):311-330.
doi: 10.1002/wrna.121 URL |
| [39] |
Vasudevan S, Tong YC, Steitz JA. Switching from repression to activation:microRNAs can up-regulate translation[J]. Science, 2007, 318(5858):1931-1934.
doi: 10.1126/science.1149460 pmid: 18048652 |
| [40] |
Murphy AJ, Guyre PM, Pioli PA. Estradiol suppresses NF-κB activation through coordinated regulation of let-7a and miR-125b in primary human macrophages[J]. J Immunol, 2010, 184(9):5029-5037.
doi: 10.4049/jimmunol.0903463 pmid: 20351193 |
| [41] |
Ørom UA, Nielsen FC, Lund AH. microRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation[J]. Mol Cell, 2008, 30(4):460-471.
doi: 10.1016/j.molcel.2008.05.001 pmid: 18498749 |
| [42] |
Guo JF, Lv J, Liu M, et al. miR-346 up-regulates argonaute 2(AGO2)protein expression to augment the activity of other microRNAs(miRNAs)and contributes to cervical cancer cell malignancy[J]. J Biol Chem, 2015, 290(51):30342-30350.
doi: 10.1074/jbc.M115.691857 URL |
| [43] |
Tsai NP, Lin YL, Wei LN. microRNA mir-346 targets the 5'-untranslated region of receptor-interacting protein 140(RIP140)mRNA and up-regulates its protein expression[J]. Biochem J, 2009, 424(3):411-418.
doi: 10.1042/BJ20090915 URL |
| [44] |
Liu MZ, Roth A, Yu M, et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis[J]. Genes Dev, 2013, 27(23):2543-2548.
doi: 10.1101/gad.224170.113 URL |
| [45] |
Xing SC, Huang CB, Wu RT, et al. Breed differences in the expression levels of gga-miR-222a in laying hens influenced H2S production by regulating methionine synthase genes in gut bacteria[J]. Microbiome, 2021, 9(1):177.
doi: 10.1186/s40168-021-01098-7 URL |
| [46] |
González-Villagra J, Kurepin LV, Reyes-Díaz MM. Evaluating the involvement and interaction of abscisic acid and miRNA156 in the induction of anthocyanin biosynthesis in drought-stressed plants[J]. Planta, 2017, 246(2):299-312.
doi: 10.1007/s00425-017-2711-y pmid: 28534253 |
| [47] |
Barrey E, Saint-Auret G, Bonnamy B, et al. Pre-microRNA and mature microRNA in human mitochondria[J]. PLoS One, 2011, 6(5):e20220.
doi: 10.1371/journal.pone.0020220 URL |
| [48] |
Bandiera S, Matégot R, Girard M, et al. MitomiRs delineating the intracellular localization of microRNAs at mitochondria[J]. Free Radic Biol Med, 2013, 64:12-19.
doi: 10.1016/j.freeradbiomed.2013.06.013 URL |
| [49] |
Bandiera S, Rüberg S, Girard M, et al. Nuclear outsourcing of RNA interference components to human mitochondria[J]. PLoS One, 2011, 6(6):e20746.
doi: 10.1371/journal.pone.0020746 URL |
| [50] |
Das S, Ferlito M, Kent OA, et al. Nuclear miRNA regulates the mitochondrial genome in the heart[J]. Circ Res, 2012, 110(12):1596-1603.
doi: 10.1161/CIRCRESAHA.112.267732 pmid: 22518031 |
| [51] |
Fan S, Tian T, Chen WX, et al. Mitochondrial miRNA determines chemoresistance by reprogramming metabolism and regulating mitochondrial transcription[J]. Cancer Res, 2019, 79(6):1069-1084.
doi: 10.1158/0008-5472.CAN-18-2505 pmid: 30659020 |
| [52] |
Zhang XR, Zuo XX, Yang B, et al. microRNA directly enhances mitochondrial translation during muscle differentiation[J]. Cell, 2014, 158(3):607-619.
doi: 10.1016/j.cell.2014.05.047 pmid: 25083871 |
| [53] |
Ro S, Ma HY, Park C, et al. The mitochondrial genome encodes abundant small noncoding RNAs[J]. Cell Res, 2013, 23(6):759-774.
doi: 10.1038/cr.2013.37 pmid: 23478297 |
| [54] |
Gusic M, Prokisch H. ncRNAs:new players in mitochondrial health and disease?[J]. Front Genet, 2020, 11:95.
doi: 10.3389/fgene.2020.00095 URL |
| [55] |
Ro S, Ma HY, Park C, et al. The mitochondrial genome encodes abundant small noncoding RNAs[J]. Cell Res, 2013, 23(6):759-774.
doi: 10.1038/cr.2013.37 pmid: 23478297 |
| [56] |
Jagannathan R, Thapa D, Nichols CE, et al. Translational regulation of the mitochondrial genome following redistribution of mitochondrial microRNA in the diabetic heart[J]. Circ Cardiovasc Genet, 2015, 8(6):785-802.
doi: 10.1161/CIRCGENETICS.115.001067 URL |
| [57] |
Vargas JNS, Kar AN, Kowalak JA, et al. Axonal localization and mitochondrial association of precursor microRNA 338[J]. Cell Mol Life Sci, 2016, 73(22):4327-4340.
pmid: 27229124 |
| [58] |
Liu JD, Valencia-Sanchez MA, Hannon GJ, et al. microRNA-dependent localization of targeted mRNAs to mammalian P-bodies[J]. Nat Cell Biol, 2005, 7(7):719-723.
pmid: 15937477 |
| [59] |
Liu JD, Rivas FV, Wohlschlegel J, et al. A role for the P-body component GW182 in microRNA function[J]. Nat Cell Biol, 2005, 7(12):1261-1266.
doi: 10.1038/ncb1333 pmid: 16284623 |
| [60] |
Toms D, Pan B, Bai YS, et al. Small RNA sequencing reveals distinct nuclear microRNAs in pig granulosa cells during ovarian follicle growth[J]. J Ovarian Res, 2021, 14(1):54.
doi: 10.1186/s13048-021-00802-3 pmid: 33879202 |
| [61] |
Vilardo E, Rossmanith W. Molecular insights into HSD10 disease:impact of SDR5C1 mutations on the human mitochondrial RNase P complex[J]. Nucleic Acids Res, 2015, 43(13):6649.
doi: 10.1093/nar/gkv658 pmid: 26092698 |
| [62] |
Metodiev MD, Thompson K, Alston CL, et al. Recessive mutations in TRMT10C cause defects in mitochondrial RNA processing and multiple respiratory chain deficiencies[J]. Am J Hum Genet, 2016, 98(5):993-1000.
doi: S0002-9297(16)30045-3 pmid: 27132592 |
| [63] | Dewe JM, Fuller BL, Lentini JM, et al. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival[J]. Mol Cell Biol, 2017, 37(21):e00214-17. |
| [64] |
Davarniya B, Hu H, Kahrizi K, et al. The role of a novel TRMT1 gene mutation and rare GRM1 gene defect in intellectual disability in two azeri families[J]. PLoS One, 2015, 10(8):e0129631.
doi: 10.1371/journal.pone.0129631 URL |
| [65] |
Najmabadi H, Hu H, Garshasbi M, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders[J]. Nature, 2011, 478(7367):57-63.
doi: 10.1038/nature10423 URL |
| [66] |
Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import[J]. Science, 2007, 315(5808):97-100.
doi: 10.1126/science.1136235 URL |
| [67] |
Matsui M, Chu YJ, Zhang HY, et al. Promoter RNA links transcriptional regulation of inflammatory pathway genes[J]. Nucleic Acids Res, 2013, 41(22):10086-10109.
doi: 10.1093/nar/gkt777 pmid: 23999091 |
| [68] |
Huang V, Place RF, Portnoy V, et al. Upregulation of Cyclin B1 by miRNA and its implications in cancer[J]. Nucleic Acids Res, 2012, 40(4):1695-1707.
doi: 10.1093/nar/gkr934 pmid: 22053081 |
| [69] |
Tang R, Li LM, Zhu DH, et al. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus:evidence for a microRNA hierarchy system[J]. Cell Res, 2012, 22(3):504-515.
doi: 10.1038/cr.2011.137 pmid: 21862971 |
| [70] | Leucci E, Patella F, Waage J, et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus[J]. Sci Reports, 2013, 3:2535. |
| [71] |
Xiao M, Li J, Li W, et al. microRNAs activate gene transcription epigenetically as an enhancer trigger[J]. RNA Biol, 2017, 14(10):1326-1334.
doi: 10.1080/15476286.2015.1112487 pmid: 26853707 |
| [72] |
Zisoulis DG, Kai ZS, Chang RK, et al. Autoregulation of microRNA biogenesis by let-7 and argonaute[J]. Nature, 2012, 486(7404):541-544.
doi: 10.1038/nature11134 URL |
| [73] |
Pu MF, Li CG, Qi XM, et al. miR-1254 suppresses HO-1 expression through seed region-dependent silencing and non-seed interaction with TFAP2A transcript to attenuate NSCLC growth[J]. PLoS Genet, 2017, 13(7):e1006896.
doi: 10.1371/journal.pgen.1006896 URL |
| [74] |
Zhao Y, Qi XM, Chen J, et al. The miR-491-3p/Sp3/ABCB1 axis attenuates multidrug resistance of hepatocellular carcinoma[J]. Cancer Lett, 2017, 408:102-111.
doi: S0304-3835(17)30514-1 pmid: 28844709 |
| [75] |
Yang XY, You CJ, Wang XF, et al. Widespread occurrence of microRNA-mediated target cleavage on membrane-bound polysomes[J]. Genome Biol, 2021, 22(1):15.
doi: 10.1186/s13059-020-02242-6 pmid: 33402203 |
| [76] |
Si WW, Li Y, Ye SY, et al. Methyltransferase 3 mediated miRNA m6A methylation promotes stress granule formation in the early stage of acute ischemic stroke[J]. Front Mol Neurosci, 2020, 13:103.
doi: 10.3389/fnmol.2020.00103 pmid: 32581712 |
| [77] |
Sharma A, Badola PK, Bhatia C, et al. Primary transcript of miR858 encodes regulatory peptide and controls flavonoid biosynthesis and development in Arabidopsis[J]. Nat Plants, 2020, 6(10):1262-1274.
doi: 10.1038/s41477-020-00769-x pmid: 32958895 |
| [78] |
Tatematsu K, Toyokura K, Okada K. Requirement of MIR165A primary transcript sequence for its activity pattern in Arabidopsis leaf primordia[J]. Plant Signal Behav, 2015, 10(8):e1055432.
doi: 10.1080/15592324.2015.1055432 URL |
| [79] |
Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28[J]. Science, 2008, 320(5872):97-100.
doi: 10.1126/science.1154040 pmid: 18292307 |
| [80] |
Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing[J]. RNA, 2008, 14(8):1539-1549.
doi: 10.1261/rna.1155108 pmid: 18566191 |
| [1] | YE Yun-fang, TIAN Qing-yin, SHI Ting-ting, WANG Liang, YUE Yuan-zheng, YANG Xiu-lian, WANG Liang-gui. Research Progress in the Biosynthesis and Regulation of β-ionone in Plants [J]. Biotechnology Bulletin, 2023, 39(8): 91-105. |
| [2] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [3] | SHI Jian-lei, ZAI Wen-shan, SU Shi-wen, FU Cun-nian, XIONG Zi-li. Identification and Expression Analysis of miRNA Related to Bacterial Wilt Resistance in Tomato [J]. Biotechnology Bulletin, 2023, 39(5): 233-242. |
| [4] | LV Yu-jing, WU Dan-dan, KONG Chun-yan, YANG Yu, GONG Ming. Genome-wide Identification of XTH Gene Family and Their Interacting miRNAs and Possible Roles in Low Temperature Adaptation in Jatropha curcas L. [J]. Biotechnology Bulletin, 2023, 39(2): 147-160. |
| [5] | YIN Guo-ying, LIU Chang, CHANG Yong-chun, YU Wang-jie, WANG Bing, ZHANG Pan, GUO Yu-shuang. Identification of the Cysteine Protease Family and Corresponding miRNAs in Nicotiana tabacum L. and Their Responses to PVY [J]. Biotechnology Bulletin, 2023, 39(10): 184-196. |
| [6] | WANG Nan-nan, WANG Wen-jia, ZHU Qiang. Research Progress of microRNAs in Plant Stress Responses [J]. Biotechnology Bulletin, 2022, 38(8): 1-11. |
| [7] | LIU Chao, CHU Hong-long, WU Li-fang, TANG Li-zhou, HAN Li-hong. Regulation Mechanism of Phosphate Homeostasis in Plants [J]. Biotechnology Bulletin, 2022, 38(2): 184-194. |
| [8] | ZHANG Chan, YAO Guang-long, ZHANG Jun-feng, YU Jing, YANG Dong-mei, CHEN Ping, WU You-gen. Research Progress on Patchoulol Molecular Regulation and Synthetic Biology in Pogostemon cablin [J]. Biotechnology Bulletin, 2021, 37(8): 55-64. |
| [9] | LI Chun-jie, WANG Cong-li. Recognition Mechanism of Plant-parasitic Nematodes in Response to Semiochemicals [J]. Biotechnology Bulletin, 2021, 37(7): 35-44. |
| [10] | CHEN Li-jie, YANG Fan, FAN Hai-yan, ZHAO Di, WANG Yuan-yuan, ZHU Xiao-feng, LIU Xiao-yu, DUAN Yu-xi. Advances of Non-coding RNA in Interactions Among Biocontrol Bacteria and Plant Nematodes and Host [J]. Biotechnology Bulletin, 2021, 37(7): 65-70. |
| [11] | ZHANG Cui-ju, MO Bei-xin, CHEN Xue-mei, CUI Jie. Advances on the Molecular Action Mechanisms of Plant miRNA [J]. Biotechnology Bulletin, 2020, 36(7): 1-14. |
| [12] | LI Ze-qing, LIU Cai-xian, XING Wen, WEN Ya-feng. Research Progress on Regulation of miRNA in the Heat Stress Response of Plants [J]. Biotechnology Bulletin, 2020, 36(2): 149-157. |
| [13] | ZHENG Wen-qing, ZHANG Qian, DU Liang. Short Tandem Target Mimic and Its Application in Analyzing Plant miRNA Functions [J]. Biotechnology Bulletin, 2020, 36(12): 256-264. |
| [14] | YANG Li-juan, LI Shi-fang, LU Mei-guang. miRNA-mediated Regulation Involved in Plant Pathogen [J]. Biotechnology Bulletin, 2020, 36(1): 101-109. |
| [15] | YAN Wu-ping, WU You-gen, YU Jing ,YANG Dong-mei, ZHANG Jun-feng. Research Progress and Prospect of microRNA in Medicinal Plants [J]. Biotechnology Bulletin, 2019, 35(8): 178-185. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||