








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (2): 90-98.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0908
Previous Articles Next Articles
ZOU Xiu-wei1( ), YUE Jia-ni1, LI Zhi-yu2, DAI Liang-ying1, LI Wei1(
), YUE Jia-ni1, LI Zhi-yu2, DAI Liang-ying1, LI Wei1( )
)
Received:2023-09-21
Online:2024-02-26
Published:2024-03-13
Contact:
LI Wei
E-mail:zouxiuwei0520@163.com;liwei350551@163.com
ZOU Xiu-wei, YUE Jia-ni, LI Zhi-yu, DAI Liang-ying, LI Wei. Functional Analysis of Rice Heat Shock Transcription Factor HsfA2b Regulating the Resistance to Abiotic Stresses[J]. Biotechnology Bulletin, 2024, 40(2): 90-98.
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| UBQ-F | AACCAGCTGAGGCCCAAGA |
| UBQ-R | ACGATTGATTTAACCAGTCCATGA |
| OsHsfA2b-F | GCCTTGTTTGATCCCCAGGA |
| OsHsfA2b-R | TTGTCGTTCAACTCCCCCTG |
| OsSOD-F | AACAAAGGCAGGGCTGTAGA |
| OsSOD-R | TAAGGTTCCAGGGCATCAAG |
| OsCAT-F | TGAAGCCAAGCATGTGAAGA |
| OsCAT-R | GCCCAACGACAACAGAAGAT |
Table 1 Primers for RT-qPCR
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| UBQ-F | AACCAGCTGAGGCCCAAGA |
| UBQ-R | ACGATTGATTTAACCAGTCCATGA |
| OsHsfA2b-F | GCCTTGTTTGATCCCCAGGA |
| OsHsfA2b-R | TTGTCGTTCAACTCCCCCTG |
| OsSOD-F | AACAAAGGCAGGGCTGTAGA |
| OsSOD-R | TAAGGTTCCAGGGCATCAAG |
| OsCAT-F | TGAAGCCAAGCATGTGAAGA |
| OsCAT-R | GCCCAACGACAACAGAAGAT |

Fig. 2 Protein structure of OsHsfA2b The number above the structural diagram represents the position of amino acids. DBD: DNA binding domain; OD: oligomerization domain; NLS: nuclear localization signal; CTAD: C-terminal transcriptional activation domain; NES: nuclear export signal

Fig. 3 Identification of OsHsfA2b-OX and OsHsfA2b-RNAi transgenic plants A: OsHsfA2b gene expression of OsHsfA2b-OX transgenic plants. B: OsHsfA2b gene expression of OsHsfA2b-RNAi transgenic plants
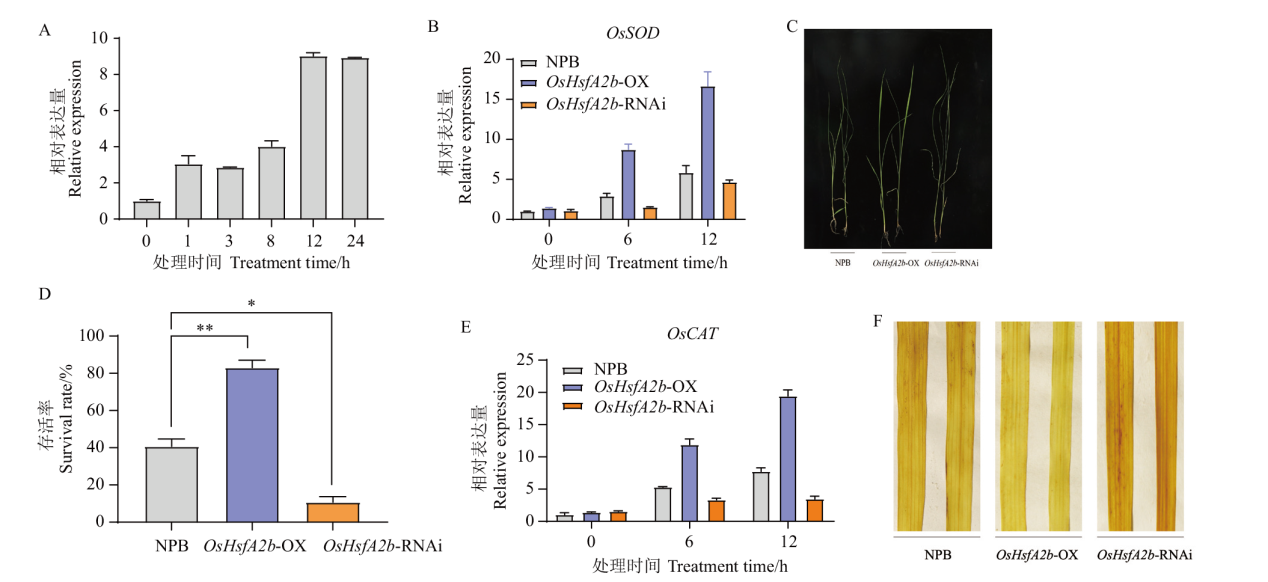
Fig. 4 Phenotypes and OsHsfA2b expressions of transgenic plants under high temperature stress A: Expression of OsHsfA2b in NPB under high temperature stress(42℃). B: Expression of OsSOD after high temperature treatment. C: Phenotypes of plants after high temperature treatment for 2 d after high temperature treatment. D: Survival rate statistic. E: Expression of OsCAT after high temperature treatment. F: Determination of reactive oxygen species content. * indicates significant difference(P<0.05), and **indicates extremely significant difference(P<0.01). The same below

Fig. 5 Phenotypes and OsHsfA2b expressions of transgenic plants under low temperature stress A: Expression of OsHsfA2b under low temperature stress(4℃). B: Phenotypes of plants after low temperature treatment for 2 d. C: Survival rate statistic. D-E: Expression of OsSOD and OsCAT after low temperature treatment

Fig. 6 Phenotypes and OsHsfA2b expressions of transgenic plants under drought stress A: Expression of OsHsfA2b under drought treatment. B: Phenotypes of plants after drought treatment for 1 d. C: Survival rate statistic. D-E: Expression of OsSOD and OsCAT after drought treatment
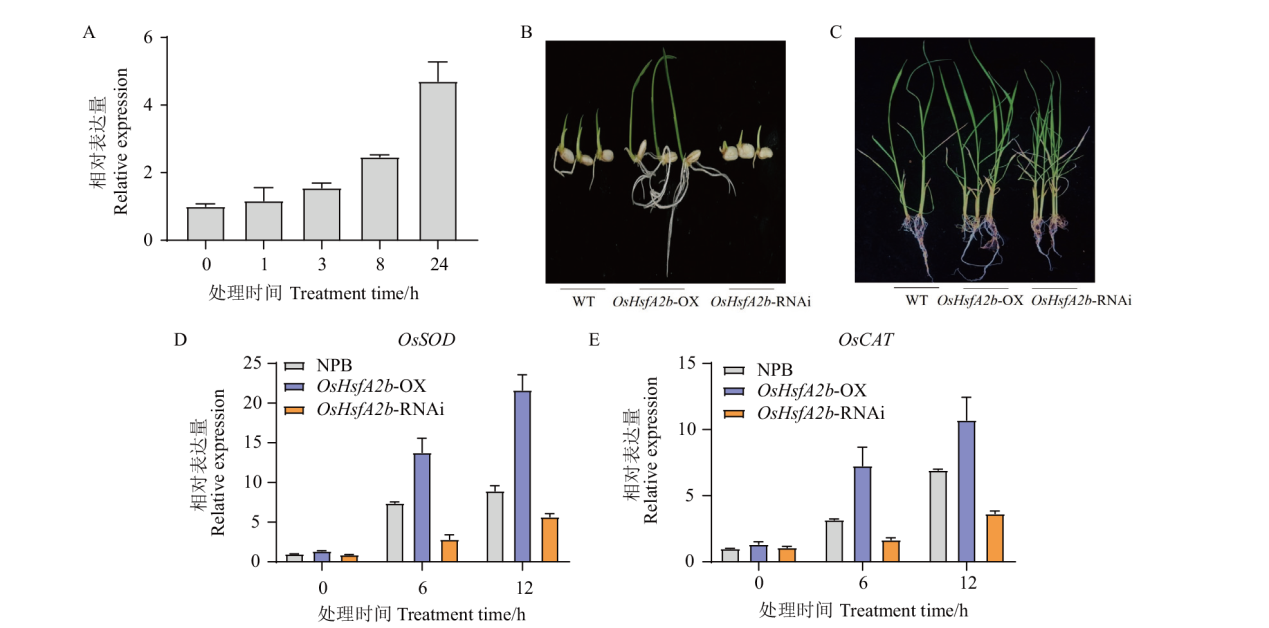
Fig. 7 Phenotypes and OsHsfA2b expressions of transgenic plants under high salt stress A: Expression of OsHsfA2b under 150 mmol/L NaCl. B: Phenotypes of seedlings geminated after 150 mmol/L NaCl for 5 d. C: Phenotypes of plants under 150 mmol/L NaCl for 2 d. D-E: Expression of OsSOD and OsCAT after high salt treatment
| [1] |
Haider S, Raza A, Iqbal J, et al. Analyzing the regulatory role of heat shock transcription factors in plant heat stress tolerance: a brief appraisal[J]. Mol Biol Rep, 2022, 49(6): 5771-5785.
doi: 10.1007/s11033-022-07190-x |
| [2] |
Ohama N, Sato H, Shinozaki K, et al. Transcriptional regulatory network of plant heat stress response[J]. Trends Plant Sci, 2017, 22(1): 53-65.
doi: S1360-1385(16)30126-1 pmid: 27666516 |
| [3] |
Li PS, Yu TF, He GH, et al. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses[J]. BMC Genomics, 2014, 15(1): 1009.
doi: 10.1186/1471-2164-15-1009 |
| [4] |
Nover L, Bharti K, Döring P, et al. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need?[J]. Cell Stress Chaperones, 2001, 6(3): 177-189.
doi: 10.1379/1466-1268(2001)006<0177:AATHST>2.0.CO;2 URL |
| [5] |
Baniwal SK, Bharti K, Chan KY, et al. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors[J]. J Biosci, 2004, 29(4): 471-487.
doi: 10.1007/BF02712120 URL |
| [6] |
Rehman A, Atif RM, Azhar MT, et al. Genome wide identification, classification and functional characterization of heat shock transcription factors in cultivated and ancestral cottons(Gossypium spp.)[J]. Int J Biol Macromol, 2021, 182: 1507-1527.
doi: 10.1016/j.ijbiomac.2021.05.016 pmid: 33965497 |
| [7] |
Lin YX, Jiang HY, Chu ZX, et al. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize[J]. BMC Genomics, 2011, 12: 76.
doi: 10.1186/1471-2164-12-76 |
| [8] | 张楠, 王映红, 王志敏, 等. 植物热激转录因子家族的研究进展[J]. 生物工程学报, 2021, 37(4): 1155-1167. |
| Zhang N, Wang YH, Wang ZM, et al. Heat shock transcription factor family in plants: a review[J]. Chin J Biotechnol, 2021, 37(4): 1155-1167. | |
| [9] | 焦淑珍, 姚文孔, 张宁波, 等. 园艺植物热激转录因子研究进展[J]. 果树学报, 2020, 37(3): 419-430. |
| Jiao SZ, Yao WK, Zhang NB, et al. Research progress of heat stress transcription factors(Hsfs)in horticultural plants[J]. J Fruit Sci, 2020, 37(3): 419-430. | |
| [10] |
Fragkostefanakis S, Mesihovic A, Simm S, et al. HsfA2 controls the activity of developmentally and stress-regulated heat stress protection mechanisms in tomato male reproductive tissues[J]. Plant Physiol, 2016, 170(4): 2461-2477.
doi: 10.1104/pp.15.01913 pmid: 26917685 |
| [11] | Zhuang L, Cao W, Wang J, et al. Characterization and functional analysis of FaHsfC1b from Festuca arundinacea conferring heat tolerance in Arabidopsis[J]. Int J Mol Sci, 2018, 19(9): E2702. |
| [12] | Guo M, Liu JH, Ma X, et al. The plant heat stress transcription factors(HSFs): Structure, regulation, and function in response to abiotic stresses[J]. Front Plant Sci, 2016, 7: 114. |
| [13] |
Chan-Schaminet KY, Baniwal SK, Bublak D, et al. Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression[J]. J Biol Chem, 2009, 284(31): 20848-20857.
doi: 10.1074/jbc.M109.007336 pmid: 19491106 |
| [14] |
Liu HC, Charng YY. Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development[J]. Plant Physiol, 2013, 163(1): 276-290.
doi: 10.1104/pp.113.221168 URL |
| [15] |
Poonia AK, Mishra SK, Sirohi P, et al. Overexpression of wheat transcription factor(TaHsfA6b)provides thermotolerance in barley[J]. Planta, 2020, 252(4): 53.
doi: 10.1007/s00425-020-03457-4 pmid: 32945950 |
| [16] |
Prieto-Dapena P, Castaño R, Almoguera C, et al. Improved resistance to controlled deterioration in transgenic seeds[J]. Plant Physiol, 2006, 142(3): 1102-1112.
doi: 10.1104/pp.106.087817 pmid: 16998084 |
| [17] |
Liu H, Zhang Y, Lu S, et al. HsfA1d promotes hypocotyl elongation under chilling via enhancing expression of ribosomal protein genes in Arabidopsis[J]. New Phytol, 2021, 231(2): 646-660.
doi: 10.1111/nph.v231.2 URL |
| [18] |
Miller G, Mittler R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants?[J]. Ann Bot, 2006, 98(2): 279-288.
doi: 10.1093/aob/mcl107 URL |
| [19] |
Pérez-Salamó I, Papdi C, Rigó G, et al. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6[J]. Plant Physiol, 2014, 165(1): 319-334.
doi: 10.1104/pp.114.237891 pmid: 24676858 |
| [20] |
Hwang SM, Kim DW, Woo MS, et al. Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions[J]. Plant Cell Environ, 2014, 37(5): 1202-1222.
doi: 10.1111/pce.2014.37.issue-5 URL |
| [21] |
Shah Z, Iqbal A, Khan FU, et al. Genetic manipulation of pea(Pisum sativum L.)with Arabidopsis's heat shock factor HsfA1d improves ROS scavenging system to confront thermal stress[J]. Genet Resour Crop Evol, 2020, 67(8): 2119-2127.
doi: 10.1007/s10722-020-00966-9 |
| [22] |
Ogawa D, Yamaguchi K, Nishiuchi T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth[J]. J Exp Bot, 2007, 58(12): 3373-3383.
doi: 10.1093/jxb/erm184 URL |
| [23] |
Mittal D, Madhyastha DA, Grover A. Gene expression analysis in response to low and high temperature and oxidative stresses in rice: combination of stresses evokes different transcriptional changes as against stresses applied individually[J]. Plant Sci, 2012, 197: 102-113.
doi: 10.1016/j.plantsci.2012.09.008 pmid: 23116677 |
| [24] | 刘爱玲. 水稻 OsHsfAs 和 OsCH2基因的功能研究[D]. 长沙: 湖南农业大学, 2010. |
| Liu AL. Study on the functions of OsHsfAs and OsCH2 genes in rice[D]. Changsha: Hunan Agricultural University, 2010. | |
| [25] |
Chauhan H, Khurana N, Agarwal P, et al. A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment[J]. PLoS One, 2013, 8(11): e79577.
doi: 10.1371/journal.pone.0079577 URL |
| [26] |
Mittler R. Oxidative stress, antioxidants and stress tolerance[J]. Trends Plant Sci, 2002, 7(9): 405-410.
doi: 10.1016/s1360-1385(02)02312-9 pmid: 12234732 |
| [27] |
Nishizawa A, Yabuta Y, Yoshida E, et al. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress[J]. Plant J, 2006, 48(4): 535-547.
doi: 10.1111/j.1365-313X.2006.02889.x pmid: 17059409 |
| [1] | WU Cui-cui, XIAO Shui-ping. Genome-wide Identification of HD-Zip Gene Family in Gossypium hirsutum L. and Expression Analysis in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(2): 130-145. |
| [2] | XIN Qi, LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua. Identification and Expression Analysis of the CBL-CIPK Gene Family in Sugarcane [J]. Biotechnology Bulletin, 2024, 40(2): 197-211. |
| [3] | LI Hao, WU Guo-qiang, WEI Ming, HAN Yue-xin. Genome-wide Identification of the BvBADH Gene Family in Sugar Beet(Beta vulgaris)and Their Expression Analysis Under High Salt Stress [J]. Biotechnology Bulletin, 2024, 40(2): 233-244. |
| [4] | LI Ya-nan, ZHANG Hao-jie, LIANG Meng-jing, LUO Tao, LI Wang-ning, ZHANG Chun-hui, JI Chun-li, LI Run-zhi, XUE Jin-ai, CUI Hong-li. Identification and Expression Analysis of Calcium-dependent Protein Kinase(CDPK)Family in Haematococcus pluvialis [J]. Biotechnology Bulletin, 2024, 40(2): 300-312. |
| [5] | ZHU Tian-yi, KONG Gui-mei, JIAO Hong-mei, GUO Ting-ting, WU Ri-han, LIU Cui-cui, GAO Cheng-feng, LI Guo-cai. Establishment of A Bacterial Model of CRISPR/Cas9 Mediated adeG Gene Knockout in Escherichia coli [J]. Biotechnology Bulletin, 2024, 40(2): 55-64. |
| [6] | WANG Jun-fang, HUANG Qiu-bin, ZHANG Piao-dan, ZHANG Peng-pai. Structure and Biosynthesis of Surfactin as well as Its Role in Biological Control [J]. Biotechnology Bulletin, 2024, 40(1): 100-112. |
| [7] | ZHANG Yi, ZHANG Xin-ru, ZHANG Jin-ke, HU Li-zong, SHANGGUAN Xin-xin, ZHENG Xiao-hong, HU Juan-juan, ZHANG Cong-cong, MU Gui-qing, LI Cheng-wei. Functional Analysis of TaMYB1 Gene in Wheat Under Cadmium Stress [J]. Biotechnology Bulletin, 2024, 40(1): 194-206. |
| [8] | BI Fang-ling, ZHAO Shuang, LI Bin, LI Ai-qin, ZHANG Jian-heng, HE Pei-min. Research Progresses and Application in the Growth-promoting Effect of Symbiotic and Epiphytic Bacteria on Green Tide-causing Ulva prolifera [J]. Biotechnology Bulletin, 2024, 40(1): 32-44. |
| [9] | LI Xue-qi, ZHANG Su-jie, YU Man, HUANG Jin-guang, ZHOU Huan-bin. Establishment of CRISPR/CasX-based Genome Editing Technology in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 40-48. |
| [10] | ZHOU Zhen-chao, ZHENG Ji, SHUAI Xin-yi, LIN Ze-jun, CHEN Hong. High-throughput Profiling and Analysis of Shared Antibiotic Resistance Genes in Human Feces, Skin and Water Environments [J]. Biotechnology Bulletin, 2023, 39(7): 288-297. |
| [11] | CHEN Yong, LI Ya-xin, WANG Ya-xuan, LIANG Lu-jie, FENG Si-yuan, Tian Guo-bao. Research Progress in the Molecular Mechanism of MCR-1 Mediated Polymyxin Resistance [J]. Biotechnology Bulletin, 2023, 39(6): 102-108. |
| [12] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [13] | LI Yuan-hong, GUO Yu-hao, CAO Yan, ZHU Zhen-zhou, WANG Fei-fei. Research Progress in the Microalgal Growth and Accumulation of Target Products Regulated by Exogenous Phytohormone [J]. Biotechnology Bulletin, 2023, 39(6): 61-72. |
| [14] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [15] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||