








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (2): 130-145.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0869
Previous Articles Next Articles
WU Cui-cui1,3( ), XIAO Shui-ping2,3(
), XIAO Shui-ping2,3( )
)
Received:2023-09-07
Online:2024-02-26
Published:2024-03-13
Contact:
XIAO Shui-ping
E-mail:wucuicui19821021@126.com;shuipingxiao@163.com
WU Cui-cui, XIAO Shui-ping. Genome-wide Identification of HD-Zip Gene Family in Gossypium hirsutum L. and Expression Analysis in Response to Abiotic Stress[J]. Biotechnology Bulletin, 2024, 40(2): 130-145.
| 基因名称Gene name | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| GhHis3 | F:CGGTGGTGTGAAGAAGCCTCAT | R:AATTTCACGAACAAGCCTCTGGAA |
| GhHDZ12 | F:CAGGCAAAGCAAAGTGGAGA | R:AGATCTGCAGCTGGTTCACT |
| GhHDZ15 | CGGGTGGGCACAAAATAAGT | TTGGTTTCAGAAATGGGGCC |
| GhHDZ46 | F:CCACCACAAAAGCCTGTTGA | R:GGTCGGAGTAATCATTGCCT |
| GhHDZ50 | F:CCAGCAAGTTTTTAGGGCGA | R:GGAACCACCGGCAAAGAATT |
| GhHDZ76 | F:ATGCTGAGCTCCAAGTCCTT | R:ATCTTGCACAACACAGCCAG |
| GhHDZ116 | F:GGCCCCATTTCTGAAACCAA | R:TGGCGACTCAACTTCTGTCT |
| GhHDZ119 | F:ACCAGCTGCAGATCTCAACT | R:CCCTTGTTGGTGTTGGTGTT |
| GhHDZ146 | F:CCACCACAAAAGCCTGTTGA | R:CGGGTCGGAGTAATCATTGT |
| GhHDZ176 | F:ATGCTGAGCTCCAAGTCCTT | R:ATCTTGCACAACACAGCCAG |
| GhHDZ188 | F:ACGTCGCACTTTTACCTTCG | R:ACCGTTTCAACCGATTCGAC |
| GhHDZ146-sub | F:TTGATACATATGCCCGTCGACATG | R:GTCGTGGTCCTTATAGTCGGATCCAT |
| GCGGGTGGGAGGGTCTTCTCTTGTAA | AAGCCGAAGACCAGAAAGCATGATCTTC |
Table 1 Primers used in the study
| 基因名称Gene name | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| GhHis3 | F:CGGTGGTGTGAAGAAGCCTCAT | R:AATTTCACGAACAAGCCTCTGGAA |
| GhHDZ12 | F:CAGGCAAAGCAAAGTGGAGA | R:AGATCTGCAGCTGGTTCACT |
| GhHDZ15 | CGGGTGGGCACAAAATAAGT | TTGGTTTCAGAAATGGGGCC |
| GhHDZ46 | F:CCACCACAAAAGCCTGTTGA | R:GGTCGGAGTAATCATTGCCT |
| GhHDZ50 | F:CCAGCAAGTTTTTAGGGCGA | R:GGAACCACCGGCAAAGAATT |
| GhHDZ76 | F:ATGCTGAGCTCCAAGTCCTT | R:ATCTTGCACAACACAGCCAG |
| GhHDZ116 | F:GGCCCCATTTCTGAAACCAA | R:TGGCGACTCAACTTCTGTCT |
| GhHDZ119 | F:ACCAGCTGCAGATCTCAACT | R:CCCTTGTTGGTGTTGGTGTT |
| GhHDZ146 | F:CCACCACAAAAGCCTGTTGA | R:CGGGTCGGAGTAATCATTGT |
| GhHDZ176 | F:ATGCTGAGCTCCAAGTCCTT | R:ATCTTGCACAACACAGCCAG |
| GhHDZ188 | F:ACGTCGCACTTTTACCTTCG | R:ACCGTTTCAACCGATTCGAC |
| GhHDZ146-sub | F:TTGATACATATGCCCGTCGACATG | R:GTCGTGGTCCTTATAGTCGGATCCAT |
| GCGGGTGGGAGGGTCTTCTCTTGTAA | AAGCCGAAGACCAGAAAGCATGATCTTC |

Fig. 3 Syntenic relationship of GhHDZ gene family A01-A13 represent the A subgroup chromosomes of upland cotton, and D01-D13 represent the D subgroup chromosomes of upland cotton
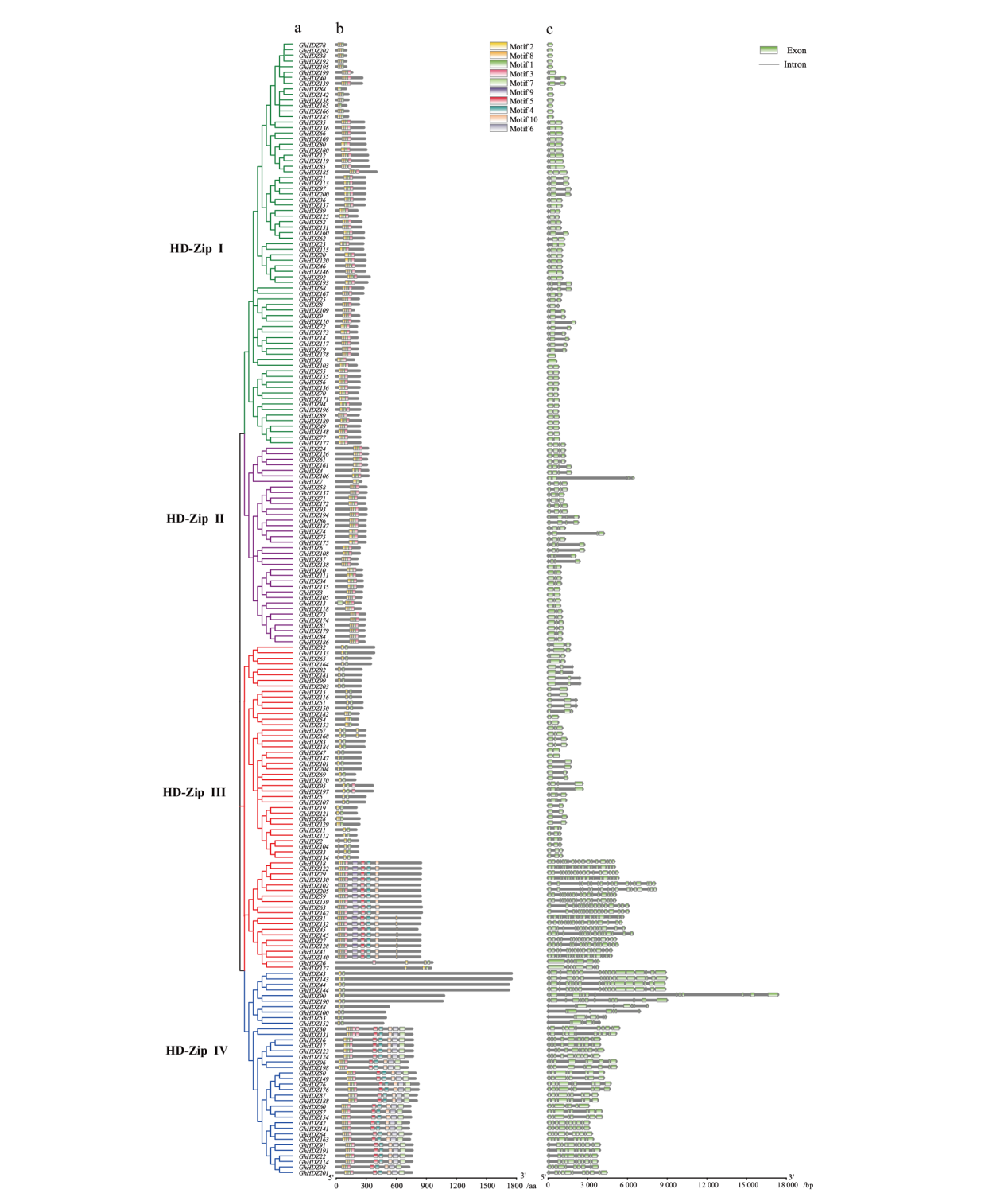
Fig. 4 Phylogenetic, gene structure, and motif analyses of GhHDZ family a: Phylogenetic relationships between GhHDZ members are based on the full-length protein sequences. b: Gene motif analysis of GhHDZ family. c: Gene structure analysis of GhHDZ family. Gene structure maps were drawn with GSDS. Green boxes indicated exons; grey lines indicated introns. The scale bar is shown at the bottom

Fig. 9 Salt stress identification of GhHDZ146 overexpression in Arabidopsis thaliana a and b: Comparison of root length between GhHDZ146-OE and WT. Bar=1 cm. c and d: Comparison of root hair length between GhHDZ146-OE and WT. Bar= 50 μm. The difference was analyzed by double-tailed t-test. The error line represents the average ±SD. Among them, WT represents wild type and OE represents GhHDZ146 overexpression strain. * P < 0.05,** P < 0.01
| [1] |
Du XM, Huang G, He SP, et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits[J]. Nat Genet, 2018, 50(6): 796-802.
doi: 10.1038/s41588-018-0116-x pmid: 29736014 |
| [2] |
Paterson AH, Wendel JF, Gundlach H, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres[J]. Nature, 2012, 492(7429): 423-427.
doi: 10.1038/nature11798 |
| [3] |
Wang KB, Wang ZW, Li FG, et al. The draft genome of a diploid cotton Gossypium raimondii[J]. Nat Genet, 2012, 44(10): 1098-1103.
doi: 10.1038/ng.2371 |
| [4] |
Yuan DJ, Tang ZH, Wang MJ, et al. The genome sequence of Sea-Island cotton(Gossypium barbadense)provides insights into the allopolyploidization and development of superior spinnable fibres[J]. Sci Rep, 2015, 5: 17662.
doi: 10.1038/srep17662 |
| [5] |
Zhang TZ, Hu Y, Jiang WK, et al. Sequencing of allotetraploid cotton(Gossypium hirsutum L. acc. TM-1)provides a resource for fiber improvement[J]. Nat Biotechnol, 2015, 33(5): 531-537.
doi: 10.1038/nbt.3207 |
| [6] |
Wang MJ, Tu LL, Yuan DJ, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense[J]. Nat Genet, 2019, 51(2): 224-229.
doi: 10.1038/s41588-018-0282-x |
| [7] |
Chen ZJ, Sreedasyam A, Ando A, et al. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement[J]. Nat Genet, 2020, 52(5): 525-533.
doi: 10.1038/s41588-020-0614-5 |
| [8] |
Huang G, Wu ZG, Percy RG, et al. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution[J]. Nat Genet, 2020, 52(5): 516-524.
doi: 10.1038/s41588-020-0607-4 pmid: 32284579 |
| [9] |
Vollbrecht E, Veit B, Sinha N, et al. The developmental gene Knotted-1 is a member of a maize homeobox gene family[J]. Nature, 1991, 350(6315): 241-243.
doi: 10.1038/350241a0 |
| [10] |
Ariel FD, Manavella PA, Dezar CA, et al. The true story of the HD-Zip family[J]. Trends Plant Sci, 2007, 12(9): 419-426.
doi: 10.1016/j.tplants.2007.08.003 pmid: 17698401 |
| [11] |
Agalou A, Purwantomo S, Overnäs E, et al. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members[J]. Plant Mol Biol, 2008, 66(1/2): 87-103.
doi: 10.1007/s11103-007-9255-7 URL |
| [12] |
Yue H, Shu DT, Wang M, et al. Genome-wide identification and expression analysis of the HD-zip gene family in wheat(Triticum aestivum L.)[J]. Genes, 2018, 9(2): 70.
doi: 10.3390/genes9020070 URL |
| [13] |
梁思维, 姜昊梁, 翟立红, 等. 玉米HD-ZIP I亚家族基因鉴定及表达分析[J]. 作物学报, 2020, 46(4): 532-543.
doi: 10.3724/SP.J.1006.2020.93040 |
|
Liang SW, Jiang HL, Zhai LH, et al. Genome-wide identification and expression analysis of HD-ZIP I subfamily genes in maize[J]. Acta Agron Sin, 2020, 46(4): 532-543.
doi: 10.3724/SP.J.1006.2020.93040 |
|
| [14] |
Li W, Dong JY, Cao MX, et al. Genome-wide identification and characterization of HD-ZIP genes in potato[J]. Gene, 2019, 697: 103-117.
doi: S0378-1119(19)30149-0 pmid: 30776460 |
| [15] |
Lin ZF, Hong YG, Yin MG, et al. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening[J]. Plant J, 2008, 55(2): 301-310.
doi: 10.1111/tpj.2008.55.issue-2 URL |
| [16] |
Manavella PA, Arce AL, Dezar CA, et al. Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb-4 transcription factor[J]. Plant J, 2006, 48(1): 125-137.
doi: 10.1111/j.1365-313X.2006.02865.x pmid: 16972869 |
| [17] |
Ariel F, Diet A, Verdenaud M, et al. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1[J]. Plant Cell, 2010, 22(7): 2171-2183.
doi: 10.1105/tpc.110.074823 URL |
| [18] |
Henriksson E, Olsson ASB, Johannesson H, et al. Homeodomain leucine zipper class I genes in Arabidopsis expression patterns and phylogenetic relationships[J]. Plant Physiol, 2005, 139(1): 509-518.
doi: 10.1104/pp.105.063461 pmid: 16055682 |
| [19] | 关淑艳, 焦鹏, 蒋振忠, 等. MYB转录因子在植物非生物胁迫中的研究进展[J]. 吉林农业大学学报, 2019, 41(3): 253-260. |
| Guan SY, Jiao P, Jiang ZZ, et al. Research progress of MYB transcription factors in plant abiotic stress[J]. J Jilin Agric Univ, 2019, 41(3): 253-260. | |
| [20] |
Himmelbach A, Hoffmann T, Leube M, et al. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis[J]. EMBO J, 2002, 21(12): 3029-3038.
pmid: 12065416 |
| [21] |
Romani F, Reinheimer R, Florent SN, et al. Evolutionary history of homeodomain leucine zipper transcription factors during plant transition to land[J]. New Phytol, 2018, 219(1): 408-421.
doi: 10.1111/nph.15133 pmid: 29635737 |
| [22] |
Valdés AE, Overnäs E, Johansson H, et al. The homeodomain-leucine zipper(HD-Zip)class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities[J]. Plant Mol Biol, 2012, 80(4/5): 405-418.
doi: 10.1007/s11103-012-9956-4 URL |
| [23] |
Zhang SX, Haider I, Kohlen W, et al. Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice[J]. Plant Mol Biol, 2012, 80(6): 571-585.
doi: 10.1007/s11103-012-9967-1 pmid: 23109182 |
| [24] |
Elhiti M, Stasolla C. Structure and function of homodomain-leucine zipper(HD-Zip)proteins[J]. Plant Signal Behav, 2009, 4(2): 86-88.
doi: 10.4161/psb.4.2.7692 URL |
| [25] |
Franklin KA, Praekelt U, Stoddart WM, et al. Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis[J]. Plant Physiol, 2003, 131(3): 1340-1346.
doi: 10.1104/pp.102.015487 pmid: 12644683 |
| [26] |
He GH, Liu P, Zhao HX, et al. The HD-ZIP II transcription factors regulate plant architecture through the auxin pathway[J]. Int J Mol Sci, 2020, 21(9): 3250.
doi: 10.3390/ijms21093250 URL |
| [27] |
Sessa G, Carabelli M, Sassi M, et al. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis[J]. Genes Dev, 2005, 19(23): 2811-2815.
doi: 10.1101/gad.364005 URL |
| [28] |
Sawa S, Ohgishi M, Goda H, et al. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis[J]. Plant J, 2002, 32(6): 1011-1022.
doi: 10.1046/j.1365-313X.2002.01488.x URL |
| [29] |
Park MY, Kim SA, Lee SJ, et al. ATHB17 is a positive regulator of abscisic acid response during early seedling growth[J]. Mol Cells, 2013, 35(2): 125-133.
doi: 10.1007/s10059-013-2245-5 pmid: 23456334 |
| [30] |
Prigge MJ, Otsuga D, Alonso JM, et al. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development[J]. Plant Cell, 2005, 17(1): 61-76.
doi: 10.1105/tpc.104.026161 URL |
| [31] |
Emery JF, Floyd SK, Alvarez J, et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes[J]. Curr Biol, 2003, 13(20): 1768-1774.
doi: 10.1016/j.cub.2003.09.035 pmid: 14561401 |
| [32] |
Baima S, Possenti M, Matteucci A, et al. The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems[J]. Plant Physiol, 2001, 126(2): 643-655.
doi: 10.1104/pp.126.2.643 pmid: 11402194 |
| [33] |
Szymanski DB, Jilk RA, Pollock SM, et al. Control of GL2 expression in Arabidopsis leaves and trichomes[J]. Development, 1998, 125(7): 1161-1171.
doi: 10.1242/dev.125.7.1161 pmid: 9477315 |
| [34] |
Peterson KM, Shyu C, Burr CA, et al. Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis[J]. Development, 2013, 140(9): 1924-1935.
doi: 10.1242/dev.090209 pmid: 23515473 |
| [35] | Mabuchi A, Soga K, Wakabayashi K, et al. Phenotypic screening of Arabidopsis T-DNA insertion lines for cell wall mechanical properties revealed ANTHOCYANINLESS2, a cell wall-related gene[J]. J Plant Physiol, 2016, 191: 29-35. |
| [36] |
Walford SA, Wu YR, Llewellyn DJ, et al. Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1[J]. Plant J, 2012, 71(3): 464-478.
doi: 10.1111/tpj.2012.71.issue-3 URL |
| [37] |
Ding MQ, Ye WW, Lin LF, et al. The hairless stem phenotype of cotton(Gossypium barbadense)is linked to a Copia-like retrotransposon insertion in a homeodomain-leucine zipper gene(HD1)[J]. Genetics, 2015, 201(1): 143-154.
doi: 10.1534/genetics.115.178236 URL |
| [38] |
Tang ML, Wu XC, Cao YF, et al. Preferential insertion of a Ty1 LTR-retrotransposon into the A sub-genome's HD1 gene significantly correlated with the reduction in stem trichomes of tetraploid cotton[J]. Mol Genet Genomics, 2020, 295(1): 47-54.
doi: 10.1007/s00438-019-01602-7 |
| [39] |
Guan XY, Pang MX, Nah G, et al. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development[J]. Nat Commun, 2014, 5: 3050.
doi: 10.1038/ncomms4050 |
| [40] |
Shan CM, Shangguan XX, Zhao B, et al. Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3[J]. Nat Commun, 2014, 5: 5519.
doi: 10.1038/ncomms6519 |
| [41] |
Lu SN, Wang JY, Chitsaz F, et al. CDD/SPARCLE: the conserved domain database in 2020[J]. Nucleic Acids Res, 2020, 48(D1): D265-D268.
doi: 10.1093/nar/gkz991 URL |
| [42] |
Gasteiger E, Gattiker A, Hoogland C, et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis[J]. Nucleic Acids Res, 2003, 31(13): 3784-3788.
doi: 10.1093/nar/gkg563 pmid: 12824418 |
| [43] |
Horton P, Park KJ, Obayashi T, et al. WoLF PSORT: protein localization predictor[J]. Nucleic Acids Res, 2007, 35(Web Server issue): W585-W587.
doi: 10.1093/nar/gkm259 pmid: 17517783 |
| [44] |
Yu CS, Chen YC, Lu CH, et al. Prediction of protein subcellular localization[J]. Proteins, 2006, 64(3): 643-651.
doi: 10.1002/prot.v64:3 URL |
| [45] |
Aiyar A. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment[J]. Methods Mol Biol, 2000, 132: 221-241.
pmid: 10547838 |
| [46] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [47] |
Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208.
doi: 10.1093/nar/gkp335 URL |
| [48] |
Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server[J]. Bioinformatics, 2015, 31(8): 1296-1297.
doi: 10.1093/bioinformatics/btu817 pmid: 25504850 |
| [49] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [50] |
Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1): 325-327.
doi: 10.1093/nar/30.1.325 pmid: 11752327 |
| [51] |
Wang YP, Tang HB, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity[J]. Nucleic Acids Res, 2012, 40(7): e49.
doi: 10.1093/nar/gkr1293 URL |
| [52] |
Wang DP, Zhang YB, Zhang Z, et al. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies[J]. Genomics Proteomics Bioinformatics, 2010, 8(1): 77-80.
doi: 10.1016/S1672-0229(10)60008-3 URL |
| [53] |
Wu CC, Xiao SP, Zuo DY, et al. Genome-wide analysis elucidates the roles of GhHMA genes in different abiotic stresses and fiber development in upland cotton[J]. Plant Physiol Biochem, 2023, 194: 281-301.
doi: 10.1016/j.plaphy.2022.11.022 URL |
| [54] |
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T)method[J]. Nat Protoc, 2008, 3(6): 1101-1108.
doi: 10.1038/nprot.2008.73 pmid: 18546601 |
| [55] |
Sparkes IA, Runions J, Kearns A, et al. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants[J]. Nat Protoc, 2006, 1(4): 2019-2025.
doi: 10.1038/nprot.2006.286 pmid: 17487191 |
| [56] |
Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana[J]. Plant J, 1998, 16(6): 735-743.
doi: 10.1046/j.1365-313x.1998.00343.x pmid: 10069079 |
| [57] | 付艳茹, 须健. 基于Image J软件的水稻根毛长度测量[J]. 湖北农业科学, 2015, 54(7): 1722-1725. |
| Fu YR, Xu J. Length measurement of the root hairs in rice based on image J software[J]. Hubei Agric Sci, 2015, 54(7): 1722-1725. | |
| [58] |
Jain M, Tyagi AK, Khurana JP. Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice[J]. FEBS J, 2008, 275(11): 2845-2861.
doi: 10.1111/j.1742-4658.2008.06424.x pmid: 18430022 |
| [59] |
Li ZQ, Zhang C, Guo YR, et al. Evolution and expression analysis reveal the potential role of the HD-Zip gene family in regulation of embryo abortion in grapes(Vitis vinifera L.)[J]. BMC Genomics, 2017, 18(1): 744.
doi: 10.1186/s12864-017-4110-y URL |
| [60] |
Chen X, Chen Z, Zhao HL, et al. Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments[J]. PLoS One, 2014, 9(2): e87156.
doi: 10.1371/journal.pone.0087156 URL |
| [61] |
Belamkar V, Weeks NT, Bharti AK, et al. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean(Glycine max)during dehydration and salt stress[J]. BMC Genomics, 2014, 15: 950.
doi: 10.1186/1471-2164-15-950 pmid: 25362847 |
| [62] |
Ding ZH, Fu LL, Yan Y, et al. Genome-wide characterization and expression profiling of HD-Zip gene family related to abiotic stress in cassava[J]. PLoS One, 2017, 12(3): e0173043.
doi: 10.1371/journal.pone.0173043 URL |
| [63] |
Shen W, Li H, Teng RM, et al. Genomic and transcriptomic analyses of HD-Zip family transcription factors and their responses to abiotic stress in tea plant(Camellia sinensis)[J]. Genomics, 2019, 111(5): 1142-1151.
doi: S0888-7543(18)30262-3 pmid: 30031053 |
| [64] |
Wei MY, Liu AL, Zhang YJ, et al. Genome-wide characterization and expression analysis of the HD-Zip gene family in response to drought and salinity stresses in sesame[J]. BMC Genomics, 2019, 20(1): 748.
doi: 10.1186/s12864-019-6091-5 pmid: 31619177 |
| [65] |
Xu GX, Guo CC, Shan HY, et al. Divergence of duplicate genes in exon-intron structure[J]. Proc Natl Acad Sci USA, 2012, 109(4): 1187-1192.
doi: 10.1073/pnas.1109047109 pmid: 22232673 |
| [66] |
Söderman E, Hjellström M, Fahleson J, et al. The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves, roots and carpels and up-regulated by water deficit conditions[J]. Plant Mol Biol, 1999, 40(6): 1073-1083.
doi: 10.1023/a:1006267013170 pmid: 10527431 |
| [67] |
Ebrahimian-Motlagh S, Ribone PA, Thirumalaikumar VP, et al. JUNGBRUNNEN1 confers drought tolerance downstream of the HD-zip I transcription factor AtHB13[J]. Front Plant Sci, 2017, 8: 2118.
doi: 10.3389/fpls.2017.02118 pmid: 29326734 |
| [1] | XIN Qi, LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua. Identification and Expression Analysis of the CBL-CIPK Gene Family in Sugarcane [J]. Biotechnology Bulletin, 2024, 40(2): 197-211. |
| [2] | LI Ya-nan, ZHANG Hao-jie, LIANG Meng-jing, LUO Tao, LI Wang-ning, ZHANG Chun-hui, JI Chun-li, LI Run-zhi, XUE Jin-ai, CUI Hong-li. Identification and Expression Analysis of Calcium-dependent Protein Kinase(CDPK)Family in Haematococcus pluvialis [J]. Biotechnology Bulletin, 2024, 40(2): 300-312. |
| [3] | ZOU Xiu-wei, YUE Jia-ni, LI Zhi-yu, DAI Liang-ying, LI Wei. Functional Analysis of Rice Heat Shock Transcription Factor HsfA2b Regulating the Resistance to Abiotic Stresses [J]. Biotechnology Bulletin, 2024, 40(2): 90-98. |
| [4] | ZHANG Yi, ZHANG Xin-ru, ZHANG Jin-ke, HU Li-zong, SHANGGUAN Xin-xin, ZHENG Xiao-hong, HU Juan-juan, ZHANG Cong-cong, MU Gui-qing, LI Cheng-wei. Functional Analysis of TaMYB1 Gene in Wheat Under Cadmium Stress [J]. Biotechnology Bulletin, 2024, 40(1): 194-206. |
| [5] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [6] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [7] | LI Yuan-hong, GUO Yu-hao, CAO Yan, ZHU Zhen-zhou, WANG Fei-fei. Research Progress in the Microalgal Growth and Accumulation of Target Products Regulated by Exogenous Phytohormone [J]. Biotechnology Bulletin, 2023, 39(6): 61-72. |
| [8] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [9] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| [10] | YANG Chun-hong, DONG Lu, CHEN Lin, SONG Li. Characterization of Soybean VAS1 Gene Family and Its Involvement in Lateral Root Development [J]. Biotechnology Bulletin, 2023, 39(3): 133-142. |
| [11] | QU Chun-juan, ZHU Yue, JIANG Chen, QU Ming-jing, WANG Xiang-yu, LI Xiao. Whole Mitochondrial Genome and Phylogeny Analysis of Anomala corpulenta [J]. Biotechnology Bulletin, 2023, 39(2): 263-273. |
| [12] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [13] | YE Hong, WANG Yu-kun. Research Progress in Immune Receptor Functions of Pattern-Recognition Receptor in Plants [J]. Biotechnology Bulletin, 2023, 39(12): 1-15. |
| [14] | REN Li, QIAO Shu-ting, GE Chen-hui, WEI Zi-tong, XU Chen-xi. Identification and Expression Analysis of Spinach PSY Gene Family [J]. Biotechnology Bulletin, 2023, 39(12): 169-178. |
| [15] | LIN Xing-yu, SONG Nan. Comparative Mitochondrial Genome and Phylogenetic Analysis of Atteva charopis Turner, 1903 [J]. Biotechnology Bulletin, 2023, 39(12): 300-310. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||