








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (2): 146-159.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0676
Previous Articles Next Articles
YANG Yan( ), HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen(
), HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen( ), ZHANG Jian-cheng(
), ZHANG Jian-cheng( )
)
Received:2023-07-14
Online:2024-02-26
Published:2024-03-13
Contact:
CHENG Chun-zhen, ZHANG Jian-cheng
E-mail:18404968975@163.com;ld0532cheng@sxau.edu.cn;zjcnd001@sxau.edu.cn
YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple[J]. Biotechnology Bulletin, 2024, 40(2): 146-159.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 引物用途 Application |
|---|---|---|---|
| MbbZIP43-F | ATGGAGCCAAATGAATCAAAGG | 53 | 基因克隆 Gene cloning |
| MbbZIP43-R | TGGCGTACTTGAGTCTTCG | ||
| MbbZIP43-OE-F | ACGGGGGACTCTAGAGGATCCATGGAGCCAAATGAATCAAAGG | 65 | 载体构建 Vector construction |
| MbbZIP43-OE-R | GCTCACCATCGCTGCACTAGTTGGCGTACTTGAGTCTTCG | ||
| ACTIN-F | TGACCGAATGAGCAAGGAAATTACT | 60 | RT-qPCR |
| ACTIN-R | TACTCAGCTTTGGCAATCCACATC | ||
| Q-MbbZIP43-F | GGAGCCAAATGAATCAAAGG | ||
| Q-MbbZIP43-R | CGCATTCTTCTTCGTCTAACT | ||
| CHS-F | GGAGACAACTGGAGAAGGACTGGAA | ||
| CHS-R | CGACATTGATACTGGTGTCTTC | ||
| CHI-F | GGGATAACCTCGCGGCCAAA | ||
| CHI-R | GCATCCATGCCGGAAGCTACAA | ||
| F3'H-F | TGGAAGCTTGTGAGGACTGGGGT | ||
| F3'H-R | CTCCTCCGATGGCAAATCAAAGA | ||
| DFR-F | CCAAGTGAAGCGGGTTGTGCT | ||
| DFR-R | CAAAGCAGGCGGACAGGAGTAGC | ||
| ANS-F | GATAGGGTTTGAGTTCAAGTA | ||
| ANS-R | TCTCCTCAGCAGCCTCAGTTTTCT | ||
| UFGT-F | CCACCGCCCTTCCAAACACTCT | ||
| UFGT-R | CACCCTTATGTTACGCGGCATGT |
Table 1 Information for the primers used in this study
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 引物用途 Application |
|---|---|---|---|
| MbbZIP43-F | ATGGAGCCAAATGAATCAAAGG | 53 | 基因克隆 Gene cloning |
| MbbZIP43-R | TGGCGTACTTGAGTCTTCG | ||
| MbbZIP43-OE-F | ACGGGGGACTCTAGAGGATCCATGGAGCCAAATGAATCAAAGG | 65 | 载体构建 Vector construction |
| MbbZIP43-OE-R | GCTCACCATCGCTGCACTAGTTGGCGTACTTGAGTCTTCG | ||
| ACTIN-F | TGACCGAATGAGCAAGGAAATTACT | 60 | RT-qPCR |
| ACTIN-R | TACTCAGCTTTGGCAATCCACATC | ||
| Q-MbbZIP43-F | GGAGCCAAATGAATCAAAGG | ||
| Q-MbbZIP43-R | CGCATTCTTCTTCGTCTAACT | ||
| CHS-F | GGAGACAACTGGAGAAGGACTGGAA | ||
| CHS-R | CGACATTGATACTGGTGTCTTC | ||
| CHI-F | GGGATAACCTCGCGGCCAAA | ||
| CHI-R | GCATCCATGCCGGAAGCTACAA | ||
| F3'H-F | TGGAAGCTTGTGAGGACTGGGGT | ||
| F3'H-R | CTCCTCCGATGGCAAATCAAAGA | ||
| DFR-F | CCAAGTGAAGCGGGTTGTGCT | ||
| DFR-R | CAAAGCAGGCGGACAGGAGTAGC | ||
| ANS-F | GATAGGGTTTGAGTTCAAGTA | ||
| ANS-R | TCTCCTCAGCAGCCTCAGTTTTCT | ||
| UFGT-F | CCACCGCCCTTCCAAACACTCT | ||
| UFGT-R | CACCCTTATGTTACGCGGCATGT |
| 软件 Software | 用途 Application | 网址 Website |
|---|---|---|
| GDR | 序列下载 | |
| CELLO v.2.5 | 亚细胞定位预测 | |
| TMHMM-2.0 | 跨膜结构预测 | |
| SignalP-5.0 | 信号肽预测 | |
| NetPhos 3.1 | 蛋白磷酸化位点预测 | |
| SOPMA | 蛋白二级结构预测 | |
| SWISS-MODEL | 蛋白三级结构预测 | |
| MEME | 蛋白保守基序预测 | |
| CDD | 结构域验证 | |
| NCBI BLASTP | 同源蛋白搜索 | |
| PlantCARE | 启动子顺式作用元件预测 | |
| TFBS | 启动子转录因子结合位点预测 | |
Table 2 Information for the software used for the bioinformatic analysis in this study
| 软件 Software | 用途 Application | 网址 Website |
|---|---|---|
| GDR | 序列下载 | |
| CELLO v.2.5 | 亚细胞定位预测 | |
| TMHMM-2.0 | 跨膜结构预测 | |
| SignalP-5.0 | 信号肽预测 | |
| NetPhos 3.1 | 蛋白磷酸化位点预测 | |
| SOPMA | 蛋白二级结构预测 | |
| SWISS-MODEL | 蛋白三级结构预测 | |
| MEME | 蛋白保守基序预测 | |
| CDD | 结构域验证 | |
| NCBI BLASTP | 同源蛋白搜索 | |
| PlantCARE | 启动子顺式作用元件预测 | |
| TFBS | 启动子转录因子结合位点预测 | |

Fig. 1 Electrophoresis detection(A)and sequence alignment(B)results of PCR products for the MbbZIP43 M: DL2000 marker; 1: using cDNA as template; 2: using gDNA as template; MbbZIP43-1: reference CDS sequence in the Malus baccata genome; MbbZIP43-2: amplified MbbZIP43 gene sequence
| 种类 Type | 功能 Function | 元件 Element | 数目 Number |
|---|---|---|---|
| 光响应元件 Light responsive elements | 光响应Light responsiveness | GATA-motif | 1 |
| 光响应Light responsiveness | MRE | 1 | |
| 光响应Light responsiveness | Box 4 | 2 | |
| 光响应Light responsiveness | ATCT-motif | 1 | |
| 光响应Light responsiveness | G-box | 1 | |
| 光响应Light responsiveness | Gap-box | 1 | |
| 光响应Light responsiveness | 3-AF1 binding site | 3 | |
| 光响应Light responsiveness | GT1-motif | 6 | |
| 生长发育相关元件 Plant growth and development related elements | 分生组织表达Meristem expression | CAT-box | 1 |
| 胚乳表达Endosperm expression | GCN4_motif | 1 | |
| 激素响应元件 Phytohormone responsive elements | 水杨酸响应Salicylic acid responsiveness | TCA-element | 1 |
| 脱落酸响应Abscisic acid responsiveness | ABRE | 1 | |
| 茉莉酸甲酯响应MeJA-responsiveness | CGTCA-motif | 2 | |
| 茉莉酸甲酯响应MeJA-responsiveness | TGACG-motif | 2 | |
| 赤霉素响应Gibberellin-responsiveness | P-box | 2 | |
| 赤霉素响应Gibberellin-responsiveness | TATC-box | 1 | |
| 逆境响应元件 Stress responsive elements | 厌氧诱导Anaerobic induction | ARE | 1 |
| 低温Low-temperature | LTR | 1 | |
| 伤害反应元件Wound-responsive element | WUN-motif | 2 | |
| 高温High-temperature | STRE | 4 | |
| 干旱响应Drought-inducibility | MYC | 4 | |
| 防御和胁迫Defense and stress | DRE core | 1 | |
| 防御和胁迫Defense and stress | TC-rich repeats | 1 | |
| 防御和胁迫Defense and stress | W box | 2 | |
| 防御和胁迫Defense and stress | MYB | 4 |
Table 3 Predicted cis-regulatory elements in the promoter region of MbbZIP43
| 种类 Type | 功能 Function | 元件 Element | 数目 Number |
|---|---|---|---|
| 光响应元件 Light responsive elements | 光响应Light responsiveness | GATA-motif | 1 |
| 光响应Light responsiveness | MRE | 1 | |
| 光响应Light responsiveness | Box 4 | 2 | |
| 光响应Light responsiveness | ATCT-motif | 1 | |
| 光响应Light responsiveness | G-box | 1 | |
| 光响应Light responsiveness | Gap-box | 1 | |
| 光响应Light responsiveness | 3-AF1 binding site | 3 | |
| 光响应Light responsiveness | GT1-motif | 6 | |
| 生长发育相关元件 Plant growth and development related elements | 分生组织表达Meristem expression | CAT-box | 1 |
| 胚乳表达Endosperm expression | GCN4_motif | 1 | |
| 激素响应元件 Phytohormone responsive elements | 水杨酸响应Salicylic acid responsiveness | TCA-element | 1 |
| 脱落酸响应Abscisic acid responsiveness | ABRE | 1 | |
| 茉莉酸甲酯响应MeJA-responsiveness | CGTCA-motif | 2 | |
| 茉莉酸甲酯响应MeJA-responsiveness | TGACG-motif | 2 | |
| 赤霉素响应Gibberellin-responsiveness | P-box | 2 | |
| 赤霉素响应Gibberellin-responsiveness | TATC-box | 1 | |
| 逆境响应元件 Stress responsive elements | 厌氧诱导Anaerobic induction | ARE | 1 |
| 低温Low-temperature | LTR | 1 | |
| 伤害反应元件Wound-responsive element | WUN-motif | 2 | |
| 高温High-temperature | STRE | 4 | |
| 干旱响应Drought-inducibility | MYC | 4 | |
| 防御和胁迫Defense and stress | DRE core | 1 | |
| 防御和胁迫Defense and stress | TC-rich repeats | 1 | |
| 防御和胁迫Defense and stress | W box | 2 | |
| 防御和胁迫Defense and stress | MYB | 4 |

Fig. 2 Multiple sequence alignment results for bZIP43 proteins and bZIP proteins related to known anthocyanin biosynthesis from different plant species MdbZIP43: Malus domestica bZIP43(XP_008393381.1); PbbZIP43: Pyrus bretschneideri bZIP43(XP_009339530.2); PybZIP43: Pyrus pyrifolia bZIP43(UXW88004.1); PabZIP43: Prunus avium bZIP43(XP_021809905.1); PsbZIP43: P. salicina bZIP43(XM_008241838.1); PdbZIP43: P. dulcis bZIP43(XP_034216145.1); PpbZIP43: P. persica bZIP43(XP_007209676.1); RcbZIP43: Rosa chinensis bZIP43(XP_024190316.2); MdbZIP44: M. domestica bZIP44(XP_008377201.2); AtHY5: Arabidopsis thaliana HY5(BAA21327.1); VvHY5: Vitis vinifera HY5(AGX85877.1); MdHY5: M. domestica HY5(NP_001280752.1); PaHY5: P. avium HY5(XP_021827650.1); PpHY5: P. persica HY5(XP_020411091.1); GbHY5: Ginkgo biloba HY5(Gb_12012). Red box, red line and green line indicates the conserved bZIP domain, α-helix and β folding, respectively

Fig. 5 Relative expressions of MbbZIP43, total phenol content, total flavonoid content, total flavonol content and anthocyanin content in ‘Hongmantang’ apple fruits at different ripening stages PS1-PS5 represent fruits at 7, 11, 15, 19 and 23 weeks post flowering, respectively. FW: Fresh weight. Different lowercase letters indicate significant differences(P<0.05). The same below
| 测定指标 Index | MbbZIP43表达水平 MbbZIP43 expression level | 总黄酮含量 Total flavonoids content | 总酚含量 Total phenolic content | 总黄酮醇含量 Total flavonol content | 花青素含量 Anthocyanin content |
|---|---|---|---|---|---|
| MbbZIP43表达水平 MbbZIP43 expression level | 1.0 | ||||
| 总黄酮含量 Total flavonoids content | 0.60 | 1.0 | |||
| 总酚含量 Total phenolic content | 0.08 | 0.69 | 1.0 | ||
| 总黄酮醇含量 Total flavonol content | -0.60 | -0.97* | -0.81 | 1.0 | |
| 花青素含量 Anthocyanin content | 0.34 | -0.02 | -0.70 | 0.21 | 1.0 |
Table 4 Correlation analysis results among the MbbZIP43 expressions, total flavonoids content, total phenolic content, total flavonol content and anthocyanin content
| 测定指标 Index | MbbZIP43表达水平 MbbZIP43 expression level | 总黄酮含量 Total flavonoids content | 总酚含量 Total phenolic content | 总黄酮醇含量 Total flavonol content | 花青素含量 Anthocyanin content |
|---|---|---|---|---|---|
| MbbZIP43表达水平 MbbZIP43 expression level | 1.0 | ||||
| 总黄酮含量 Total flavonoids content | 0.60 | 1.0 | |||
| 总酚含量 Total phenolic content | 0.08 | 0.69 | 1.0 | ||
| 总黄酮醇含量 Total flavonol content | -0.60 | -0.97* | -0.81 | 1.0 | |
| 花青素含量 Anthocyanin content | 0.34 | -0.02 | -0.70 | 0.21 | 1.0 |
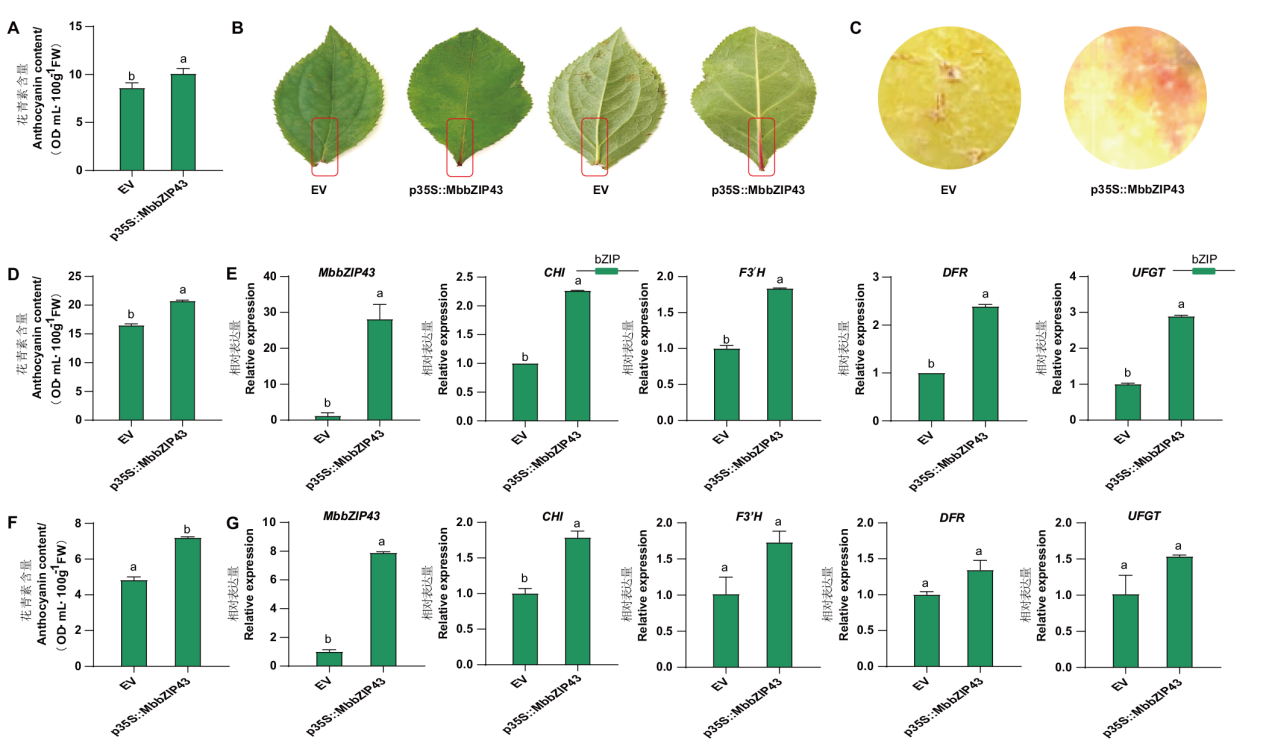
Fig. 6 Influences of transient overexpression of MbbZIP43 gene on anthocyanin accumulations A: Influences of transient overexpression of MbbZIP43 gene on the anthocyanin accumulation in tobacco. B: Apple leaves overexpressing MbbZIP43. C: Apple peel overexpressing MbbZIP43. D: Influences of transient overexpression of MbbZIP43 gene on anthocyanin accumulation in apple leaves. E: Influences of transient overexpression of MbbZIP43 gene on the expressions of MbbZIP43 and structural genes related to anthocyanin biosynthesis in apple leaves. F: Influences of transient overexpression of MbbZIP43 gene on anthocyanin accumulation in apple. G: Influences of transient overexpression of MbbZIP43 gene on the expressions of MbbZIP43 and structural genes related to anthocyanin biosynthesis in apple peel
| [1] |
Hurst HC. Transcription factors 1: bZIP proteins[J]. Protein Profile, 1995, 2(2): 101-168.
pmid: 7780801 |
| [2] |
Ellenberger T. Getting a grip on DNA recognition: structures of the basic region leucine zipper, and the basic region helix-loop-helix DNA-binding domains[J]. Curr Opin Struct Biol, 1994, 4(1): 12-21.
doi: 10.1016/S0959-440X(94)90054-X URL |
| [3] |
Wei KF, Chen J, et al. Genome-wide analysis of bZIP-encoding genes in maize[J]. DNA Res, 2012, 19(6): 463-476.
doi: 10.1093/dnares/dss026 pmid: 23103471 |
| [4] |
Silveira AB, Gauer L, Tomaz JP, et al. The Arabidopsis AtbZIP9 protein fused to the VP16 transcriptional activation domain alters leaf and vascular development[J]. Plant Sci, 2007, 172(6): 1148-1156.
doi: 10.1016/j.plantsci.2007.03.003 URL |
| [5] |
Yamamoto MP, Onodera Y, Touno SM, et al. Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes[J]. Plant Physiol, 2006, 141(4): 1694-1707.
doi: 10.1104/pp.106.082826 pmid: 16798940 |
| [6] |
Burman N, Bhatnagar A, Khurana JP. OsbZIP48, a HY5 transcription factor ortholog, exerts pleiotropic effects in light-regulated development[J]. Plant Physiol, 2018, 176(2): 1262-1285.
doi: 10.1104/pp.17.00478 pmid: 28775143 |
| [7] |
Babu Rajendra Prasad V, Gupta N, Nandi A, et al. HY1 genetically interacts with GBF1 and regulates the activity of the Z-box containing promoters in light signaling pathways in Arabidopsis thaliana[J]. Mech Dev, 2012, 129(9/10/11/12): 298-307.
doi: 10.1016/j.mod.2012.06.004 URL |
| [8] |
Yang SS, Zhang XX, Zhang XM, et al. A bZIP transcription factor, PqbZIP1, is involved in the plant defense response of American ginseng[J]. PeerJ, 2022, 10: e12939.
doi: 10.7717/peerj.12939 URL |
| [9] |
Collin A, Daszkowska-Golec A, Szarejko I. Updates on the role of abscisic acid insensitive 5(abi5)and abscisic acid-responsive element binding factors(abfs)in ABA signaling in different developmental stages in plants[J]. Cells, 2021, 10(8): 1996.
doi: 10.3390/cells10081996 URL |
| [10] |
Alves M, Dadalto S, Gonçalves A, et al. Plant bZIP transcription factors responsive to pathogens: a review[J]. Int J Mol Sci, 2013, 14(4): 7815-7828.
doi: 10.3390/ijms14047815 pmid: 23574941 |
| [11] |
刘恺媛, 王茂良, 辛海波, 等. 植物花青素合成与调控研究进展[J]. 中国农学通报, 2021, 37(14): 41-51.
doi: 10.11924/j.issn.1000-6850.casb2020-0390 |
|
Liu KY, Wang ML, Xin HB, et al. Anthocyanin biosynthesis and regulate mechanisms in plants: a review[J]. Chin Agric Sci Bull, 2021, 37(14): 41-51.
doi: 10.11924/j.issn.1000-6850.casb2020-0390 |
|
| [12] |
Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis[J]. Trends Plant Sci, 2010, 15(10): 573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [13] |
Shin DH, Choi M, Kim K, et al. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis[J]. FEBS Lett, 2013, 587(10): 1543-1547.
doi: 10.1016/j.febslet.2013.03.037 URL |
| [14] |
Zhao Y, Min T, et al. The photomorphogenic transcription factor PpHY5 regulates anthocyanin accumulation in response to UVA and UVB irradiation[J]. Front Plant Sci, 2021, 11: 603178.
doi: 10.3389/fpls.2020.603178 URL |
| [15] |
Wang YY, Zhang XD, Zhao YR, et al. Transcription factor PyHY5 binds to the promoters of PyWD40 and PyMYB10 and regulates its expression in red pear ‘Yunhongli No. 1’[J]. Plant Physiol Biochem, 2020, 154: 665-674.
doi: 10.1016/j.plaphy.2020.07.008 URL |
| [16] |
Liu YQ, Ye YT, et al. B-box transcription factor FaBBX22 promotes light-induced anthocyanin accumulation in strawberry(Fragar-ia×ananassa)[J]. Int J Mol Sci, 2022, 23(14): 7757.
doi: 10.3390/ijms23147757 URL |
| [17] |
Liu YQ, Tang L, Wang YP, et al. The blue light signal transduction module FaCRY1-FaCOP1-FaHY5 regulates anthocyanin accumulation in cultivated strawberry[J]. Front Plant Sci, 2023, 14: 1144273.
doi: 10.3389/fpls.2023.1144273 URL |
| [18] |
Liu HN, Su J, Zhu YF, et al. The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter[J]. Hortic Res, 2019, 6: 134.
doi: 10.1038/s41438-019-0217-4 |
| [19] |
Xu ZJ, Wang JC, Ma YB, et al. The bZIP transcription factor SlAREB1 regulates anthocyanin biosynthesis in response to low temperature in tomato[J]. Plant J, 2023, 115(1): 205-219.
doi: 10.1111/tpj.v115.1 URL |
| [20] |
An JP, Yao JF, et al. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation[J]. Plant Cell Environ, 2018, 41(11): 2678-2692.
doi: 10.1111/pce.v41.11 URL |
| [21] |
An JP, Zhang XW, Liu YJ, et al. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple[J]. J Exp Bot, 2021, 72(4): 1460-1472.
doi: 10.1093/jxb/eraa525 URL |
| [22] |
Liu WJ, Mei ZX, Yu L, et al. The ABA-induced NAC transcription factor MdNAC1 interacts with a bZIP-type transcription factor to promote anthocyanin synthesis in red-fleshed apples[J]. Hortic Res, 2023, 10(5): uhad049.
doi: 10.1093/hr/uhad049 URL |
| [23] | 杨廷桢, 高敬东, 王骞, 等. 苹果属观赏新品种——‘红满堂’的选育[J]. 果树学报, 2015, 32(4): 727-729, 520. |
| Yang TZ, Gao JD, Wang Q, et al. A new Malus ornamental variety ‘Hongmantang’[J]. J Fruit Sci, 2015, 32(4): 727-729, 520. | |
| [24] | 郭子微, 侯文赫, 付鸿博, 等. 不同苹果果实发育过程中酚类物质含量及抗氧化能力变化研究[J]. 山东农业科学, 2021, 53(11): 35-44. |
| Guo ZW, Hou WH, Fu HB, et al. Changes of phenolic substances and antioxidant capacity during fruit development of different apple varieties[J]. Shandong Agric Sci, 2021, 53(11): 35-44. | |
| [25] |
Cheng CZ, Guo ZW, Li HA, et al. Integrated metabolic, transcriptomic and chromatin accessibility analyses provide novel insights into the competition for anthocyanins and flavonols biosynthesis during fruit ripening in red apple[J]. Front Plant Sci, 2022, 13: 975356.
doi: 10.3389/fpls.2022.975356 URL |
| [26] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [27] | 刘嘉鹏, 武欢, 王斌, 等. 香蕉MaSNAT2基因的克隆与表达分析[J]. 西北植物学报, 2022, 42(4): 569-577. |
| Liu JP, Wu H, Wang B, et al. Cloning and expression analysis of a MaSNAT2 gene in banana[J]. Acta Bot Boreali Occidentalia Sin, 2022, 42(4): 569-577. | |
| [28] |
Cheng CZ, Zhong Y, et al. The upregulated expression of the Citrus RIN4 gene in HLB diseased Citrus aids Candidatus Liberibacter asiaticus infection[J]. Int J Mol Sci, 2022, 23(13): 6971.
doi: 10.3390/ijms23136971 URL |
| [29] |
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method[J]. Nat Protoc, 2008, 3(6): 1101-1108.
doi: 10.1038/nprot.2008.73 pmid: 18546601 |
| [30] |
Slinkard K, Singleton VL. Total phenol analysis: automation and comparison with manual methods[J]. Am J Enol Vitic, 1977, 28(1): 49-55.
doi: 10.5344/ajev.1977.28.1.49 URL |
| [31] |
Zheng HZ, Kim YI, Chung SK. A profile of physicochemical and antioxidant changes during fruit growth for the utilisation of unripe apples[J]. Food Chem, 2012, 131(1): 106-110.
doi: 10.1016/j.foodchem.2011.08.038 URL |
| [32] | 仝月澳, 周厚基. 果树营养诊断法[M]. 北京: 农业出版社, 1982. |
| Tong YA, Zhou HJ. Nutritional diagnosis of fruit trees[M]. Beijing: Agricultural Publishing House, 1982. | |
| [33] |
Wang B, Xu YB, Xu SY, et al. Characterization of banana SNARE genes and their expression analysis under temperature stress and mutualistic and pathogenic fungal colonization[J]. Plants, 2023, 12(8): 1599.
doi: 10.3390/plants12081599 URL |
| [34] |
Alabd A, Ahmad M, Zhang X, et al. Light-responsive transcription factor PpWRKY44 induces anthocyanin accumulation by regulating PpMYB10 expression in pear[J]. Hortic Res, 2022, 9: uhac199.
doi: 10.1093/hr/uhac199 URL |
| [35] |
An JP, Wang XF, Li YY, et al. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation[J]. Plant Physiol, 2018, 178(2): 808-823.
doi: 10.1104/pp.18.00068 URL |
| [36] |
Yu LJ, Sun YY, et al. ROS1 promotes low temperature-induced anthocyanin accumulation in apple by demethylating the promoter of anthocyanin-associated genes[J]. Hortic Res, 2022, 9: uhac007.
doi: 10.1093/hr/uhac007 URL |
| [37] | Zhang YY, Huang DQ, Wang B, et al. Characterization of highbush blueberry(Vaccinium corymbosum L.)anthocyanin biosynthesis related MYBs and functional analysis of VcMYB gene[J]. Curr News Mol Biol, 2023, 45(1): 379-399. |
| [38] |
Zhang YY, Liu F, Wang B, et al. Identification, characterization and expression analysis of anthocyanin biosynthesis-related bHLH genes in blueberry(Vaccinium corymbosum L.)[J]. Int J Mol Sci, 2021, 22(24): 13274.
doi: 10.3390/ijms222413274 URL |
| [39] |
Takos AM, Jaffé FW, Jacob SR, et al. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples[J]. Plant Physiol, 2006, 142(3): 1216-1232.
doi: 10.1104/pp.106.088104 pmid: 17012405 |
| [40] |
Telias A, Kui LW, Stevenson DE, et al. Apple skin patterning is associated with differential expression of MYB10[J]. BMC Plant Biol, 2011, 11: 93.
doi: 10.1186/1471-2229-11-93 pmid: 21599973 |
| [41] |
Vimolmangkang S, Han YP, Wei GC, et al. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development[J]. BMC Plant Biol, 2013, 13: 176.
doi: 10.1186/1471-2229-13-176 pmid: 24199943 |
| [42] |
An JP, Li HH, Song LQ, et al. The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple[J]. Plant Physiol Biochem, 2016, 108: 24-31.
doi: 10.1016/j.plaphy.2016.06.032 URL |
| [43] |
An XH, Tian Y, Chen KQ, et al. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation[J]. J Plant Physiol, 2012, 169(7): 710-717.
doi: 10.1016/j.jplph.2012.01.015 URL |
| [44] |
An JP, Qu FJ, Yao JF, et al. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple[J]. Hortic Res, 2017, 4: 17023.
doi: 10.1038/hortres.2017.23 URL |
| [45] |
Li XL, Cheng YD, et al. Weighted gene coexpression correlation network analysis reveals a potential molecular regulatory mechanism of anthocyanin accumulation under different storage temperatures in ‘Friar’ plum[J]. BMC Plant Biol, 2021, 21(1): 576.
doi: 10.1186/s12870-021-03354-2 |
| [46] |
Hu JF, Fang HC, Wang J, et al. Ultraviolet B-induced MdWRKY72 expression promotes anthocyanin synthesis in apple[J]. Plant Sci, 2020, 292: 110377.
doi: 10.1016/j.plantsci.2019.110377 URL |
| [47] |
An JP, Wang XF, et al. MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation[J]. Plant Biotechnol J, 2019, 17(12): 2231-2233.
doi: 10.1111/pbi.v17.12 URL |
| [48] |
Rudell DR, Mattheis JP. Synergism exists between ethylene and methyl jasmonate in artificial light-induced pigment enhancement of ‘Fuji’ apple fruit peel[J]. Postharvest Biol Technol, 2008, 47(1): 136-140.
doi: 10.1016/j.postharvbio.2007.05.021 URL |
| [49] |
Sun JJ, Wang YC, Chen XS, et al. Effects of methyl jasmonate and abscisic acid on anthocyanin biosynthesis in callus cultures of red-fleshed apple(Malus sieversii f. niedzwetzkyana)[J]. Plant Cell Tiss Organ Cult, 2017, 130(2): 227-237.
doi: 10.1007/s11240-017-1217-4 URL |
| [50] |
Chen ZJ, Yu L, Liu WJ, et al. Research progress of fruit color development in apple(Malus domestica Borkh.)[J]. Plant Physiol Biochem, 2021, 162: 267-279.
doi: 10.1016/j.plaphy.2021.02.033 URL |
| [51] |
Ubi BE, Honda C, Bessho H, et al. Expression analysis of anthocyanin biosynthetic genes in apple skin: effect of UV-B and temperature[J]. Plant Sci, 2006, 170(3): 571-578.
doi: 10.1016/j.plantsci.2005.10.009 URL |
| [52] |
Kui LW, Micheletti D, Palmer J, et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex[J]. Plant Cell Environ, 2011, 34(7): 1176-1190.
doi: 10.1111/pce.2011.34.issue-7 URL |
| [53] |
An JP, Zhang XW, Bi SQ, et al. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple[J]. Plant J, 2020, 101(3): 573-589.
doi: 10.1111/tpj.v101.3 URL |
| [54] |
An JP, Zhang XW, You CX, et al. MdWRKY40 promotes wounding-induced anthocyanin biosynthesis in association with MdMYB1 and undergoes MdBT2-mediated degradation[J]. New Phytol, 2019, 224(1): 380-395.
doi: 10.1111/nph.v224.1 URL |
| [55] | 杜婷婷, 宋治华, 董碧莹, 等. 木豆类黄酮代谢通路关键基因家族的鉴定与表达分析[J]. 农业生物技术学报, 2021, 29(12): 2289-2303. |
| Du TT, Song ZH, Dong BY, et al. Identification and expression analysis of key gene families in flavonoid metabolism pathway in pigeon pea(Cajanus cajan)[J]. J Agric Biotechnol, 2021, 29(12): 2289-2303. | |
| [56] |
Meng JX, Yin J, Wang H, et al. A TCP transcription factor in Malus halliana, MhTCP4, positively regulates anthocyanins biosynthesis[J]. Int J Mol Sci, 2022, 23(16): 9051.
doi: 10.3390/ijms23169051 URL |
| [1] | REN Yan-jing, ZHANG Lu-gang, ZHAO Meng-liang, LI Jiang, SHAO Deng-kui. cDNA Yeast Library Construction of Chinese Cabbage Seeds and Screening and Analysis of BrTTG1 Interacting Proteins [J]. Biotechnology Bulletin, 2024, 40(2): 223-232. |
| [2] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| [3] | XIE Hong, ZHOU Li-ying, LI Shu-wen, WANG Meng-di, AI Ye, CHAO Yue-hui. Structural and Functional Analysis of MtCIM Gene in Medicago truncatula [J]. Biotechnology Bulletin, 2024, 40(1): 262-269. |
| [4] | TANG Wei-lin, KANG Qin, WANG Xia, SHEN Ming-yang, SUN Xin-jiang, WANG Ke, HOU Kai, WU Wei, XU Dong-bei. Cloning and Expression Pattern Analysis of Jasmonic Acid Receptor Gene McCOI1a in Mentha canadensis L. [J]. Biotechnology Bulletin, 2024, 40(1): 270-280. |
| [5] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [6] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [7] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [8] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [9] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [10] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [11] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [12] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [13] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [14] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [15] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||