








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (4): 52-66.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1044
Previous Articles Next Articles
YUAN Xiao-yan1,2( ), ZHANG Yu-ge1,2, YAO Li-juan1,2, TANG Chen3, LI Xiao-juan1,2(
), ZHANG Yu-ge1,2, YAO Li-juan1,2, TANG Chen3, LI Xiao-juan1,2( )
)
Received:2023-11-07
Online:2024-04-26
Published:2024-04-30
Contact:
LI Xiao-juan
E-mail:yuanxy@bjfu.edu.cn;lixj@bjfu.edu.cn
YUAN Xiao-yan, ZHANG Yu-ge, YAO Li-juan, TANG Chen, LI Xiao-juan. Application of Single-virus Tracking Technique in Animal and Plant Cells[J]. Biotechnology Bulletin, 2024, 40(4): 52-66.

Fig. 1 Life cycles of animal virus and plant virus After a series of steps such as receptor contact, entry, transport, uncoating, replication, translation and assembly, the animal virus synthesized the progeny virus and left the cell through exocytosis, budding or lysis. Plant viruses undergo a similar process to animal viruses after entering the cells. The difference is that they leave the primary infected cells through the plant plasmodesmata and carry out intercellular transport after completing the assembly of progeny viruses

Fig. 2 Virus fluorescence labeling strategies The commonly used fluorescence labeling strategies in single virus tracking technology can be divided into three categories: Chemical labeling, coding tag labeling and encoding fluorescent protein labeling. Chemical labeling methods include cross-linking reaction, click chemistry and biotin-streptavidin interaction; coding tag labeling methods include peptide tag-mediated labeling and oligonucleotide-guided labeling
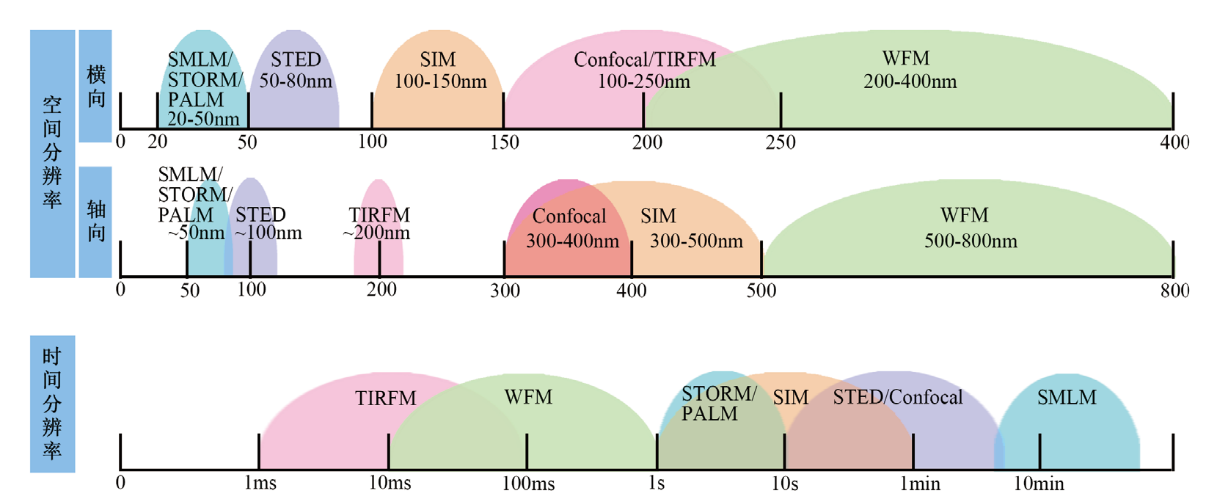
Fig. 3 Spatiotemporal resolution of different imaging techniques The figure shows, from top to bottom, the range of spatial and temporal resolution of the fluorescent microscopy imaging techniques commonly used in single-virus tracking. Refer to Schermelleh et al[125]
| [1] |
Pellett PE, Mitra S, Holland TC. Basics of virology[J]. Handb Clin Neurol, 2014, 123: 45-66.
doi: 10.1016/B978-0-444-53488-0.00002-X pmid: 25015480 |
| [2] |
Simmonds P, Aiewsakun P. Virus classification - where do you draw the line?[J]. Arch Virol, 2018, 163(8): 2037-2046.
doi: 10.1007/s00705-018-3938-z pmid: 30039318 |
| [3] |
Greber UF, Way M. A superhighway to virus infection[J]. Cell, 2006, 124(4): 741-754.
doi: 10.1016/j.cell.2006.02.018 pmid: 16497585 |
| [4] |
Döhner K, Nagel CH, Sodeik B. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins[J]. Trends Microbiol, 2005, 13(7): 320-327.
doi: 10.1016/j.tim.2005.05.010 pmid: 15950476 |
| [5] |
李家俊, 郑潇, 盛杰, 等. 新型冠状病毒及其临床检测方法研究进展[J]. 生物技术通报, 2021, 37(4): 282-292.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0954 |
| Li JJ, Zheng X, Sheng J, et al. Novel coronavirus and research progress of related clinical detection methods[J]. Biotechnol Bull, 2021, 37(4): 282-292. | |
| [6] |
Diakou KI, Mitsis T, Pierouli K, et al. Ebola virus disease and current therapeutic strategies: a review[J]. Adv Exp Med Biol, 2021, 1339: 131-137.
doi: 10.1007/978-3-030-78787-5_18 pmid: 35023100 |
| [7] |
Li YJ, Mohammadi A, Li JZ. Challenges and promise of human immunodeficiency virus remission[J]. J Infect Dis, 2021, 223(12Suppl 2): 4-12.
doi: 10.1093/infdis/jiaa567 URL |
| [8] |
Musso D, Gubler DJ. Zika virus[J]. Clin Microbiol Rev, 2016, 29(3): 487-524.
doi: 10.1128/CMR.00072-15 pmid: 27029595 |
| [9] |
Jeng WJ, Papatheodoridis GV, Lok ASF. Hepatitis B[J]. Lancet, 2023, 401(10381): 1039-1052.
doi: 10.1016/S0140-6736(22)01468-4 URL |
| [10] |
Creager ANH. Tobacco mosaic virus and the history of molecular biology[J]. Annu Rev Virol, 2022, 9(1): 39-55.
doi: 10.1146/annurev-virology-100520-014520 pmid: 35704746 |
| [11] |
邱艳红, 王超楠, 朱水芳. 黄瓜花叶病毒致病性研究进展[J]. 生物技术通报, 2017, 33(9): 10-16.
doi: 10.13560/j.cnki.biotech.bull.1985.2017-0278 |
| Qiu YH, Wang CN, Zhu SF. Research advances on the pathogenicity of cucumber mosaic virus[J]. Biotechnol Bull, 2017, 33(9): 10-16. | |
| [12] | Qi SM, Zhang SJ, Islam MM, et al. Natural resources resistance to tomato spotted wilt virus(TSWV)in tomato(Solanum lycopersicum)[J]. Int J Mol Sci, 2021, 22(20): 10978. |
| [13] |
Bragard C, Caciagli P, Lemaire O, et al. Status and prospects of plant virus control through interference with vector transmission[J]. Annu Rev Phytopathol, 2013, 51: 177-201.
doi: 10.1146/annurev-phyto-082712-102346 pmid: 23663003 |
| [14] |
Jones RAC. Plant virus emergence and evolution: origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control[J]. Virus Res, 2009, 141(2): 113-130.
doi: 10.1016/j.virusres.2008.07.028 pmid: 19159652 |
| [15] |
Nakai M, Goto T. Ultrastructure and morphogenesis of human immunodeficiency virus[J]. J Electron Microsc, 1996, 45(4): 247-257.
pmid: 8888583 |
| [16] |
Zhang YB, Wang YJ, Xie ZK, et al. Purification and immuno-gold labeling of lily mottle virus from lily leaves[J]. J Virol Methods, 2016, 232: 33-38.
doi: 10.1016/j.jviromet.2016.02.001 pmid: 26927705 |
| [17] |
Brookes SM, Hyatt AD, Eaton BT. The use of immuno-gold silver staining in bluetongue virus adsorption and neutralisation studies[J]. J Virol Methods, 1994, 46(2): 117-132.
pmid: 8188809 |
| [18] |
Timpe J, Bevington J, Casper J, et al. Mechanisms of adeno-associated virus genome encapsidation[J]. Curr Gene Ther, 2005, 5(3): 273-284.
pmid: 15975005 |
| [19] |
Smith AE, Helenius A. How viruses enter animal cells[J]. Science, 2004, 304(5668): 237-242.
doi: 10.1126/science.1094823 pmid: 15073366 |
| [20] |
Henry T, Gorvel JP, Méresse S. Molecular motors hijacking by intracellular pathogens[J]. Cell Microbiol, 2006, 8(1): 23-32.
doi: 10.1111/j.1462-5822.2005.00649.x pmid: 16367863 |
| [21] |
Zhou A, Zhang WH, Dong X, et al. The battle for autophagy between host and influenza A virus[J]. Virulence, 2022, 13(1): 46-59.
doi: 10.1080/21505594.2021.2014680 pmid: 34967267 |
| [22] |
Cruz-Oliveira C, Freire JM, Conceição TM, et al. Receptors and routes of dengue virus entry into the host cells[J]. FEMS Microbiol Rev, 2015, 39(2): 155-170.
doi: 10.1093/femsre/fuu004 pmid: 25725010 |
| [23] |
Rust MJ, Lakadamyali M, Zhang F, et al. Assembly of endocytic machinery around individual influenza viruses during viral entry[J]. Nat Struct Mol Biol, 2004, 11(6): 567-573.
doi: 10.1038/nsmb769 pmid: 15122347 |
| [24] |
Bayati A, Kumar R, Francis V, et al. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis[J]. J Biol Chem, 2021, 296: 100306.
doi: 10.1016/j.jbc.2021.100306 URL |
| [25] |
Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin[J]. Nat Rev Microbiol, 2006, 4(1): 67-76.
pmid: 16357862 |
| [26] |
Li Q, Li W, Yin W, et al. Single-particle tracking of human immunodeficiency virus type 1 productive entry into human primary macrophages[J]. ACS Nano, 2017, 11(4): 3890-3903.
doi: 10.1021/acsnano.7b00275 pmid: 28371581 |
| [27] |
Liu SL, Zhang ZL, Tian ZQ, et al. Effectively and efficiently dissecting the infection of influenza virus by quantum-dot-based single-particle tracking[J]. ACS Nano, 2012, 6(1): 141-150.
doi: 10.1021/nn2031353 URL |
| [28] | Owusu IA, Quaye O, Passalacqua KD, et al. Egress of non-enveloped enteric RNA viruses[J]. J Gen Virol, 2021, 102(3): 001557. |
| [29] |
Calistri A, Salata C, Parolin C, et al. Role of multivesicular bodies and their components in the egress of enveloped RNA viruses[J]. Rev Med Virol, 2009, 19(1): 31-45.
doi: 10.1002/rmv.588 pmid: 18618839 |
| [30] | Yuan C, Lazarowitz SG, Citovsky V. The plasmodesmal localization signal of TMV MP is recognized by plant synaptotagmin SYTA[J]. mBio, 2018, 9(4): e01314-e01318. |
| [31] |
Hong JS, Ju HJ. The plant cellular systems for plant virus movement[J]. Plant Pathol J, 2017, 33(3): 213-228.
doi: 10.5423/PPJ.RW.09.2016.0198 URL |
| [32] |
Liu SL, Wang ZG, Xie HY, et al. Single-virus tracking: from imaging methodologies to virological applications[J]. Chem Rev, 2020, 120(3): 1936-1979.
doi: 10.1021/acs.chemrev.9b00692 URL |
| [33] |
王志刚, 刘书琳, 刘安安, 等. 单病毒示踪[J]. 化学进展, 2021, 33(1): 13-24.
doi: 10.7536/PC201045 |
| Wang ZG, Liu SL, Liu AA, et al. Single-virus tracking[J]. Prog Chem, 2021, 33(1): 13-24. | |
| [34] |
Wagh SB, Maslivetc VA, La Clair JJ, et al. Lessons in organic fluorescent probe discovery[J]. Chembiochem, 2021, 22(22): 3109-3139.
doi: 10.1002/cbic.202100171 pmid: 34062039 |
| [35] |
Li XC, Liang X, Yin JL, et al. Organic fluorescent probes for monitoring autophagy in living cells[J]. Chem Soc Rev, 2021, 50(1): 102-119.
doi: 10.1039/d0cs00896f pmid: 33155002 |
| [36] |
Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, et al. Quantum dots versus organic dyes as fluorescent labels[J]. Nat Methods, 2008, 5(9): 763-775.
doi: 10.1038/nmeth.1248 pmid: 18756197 |
| [37] |
Huang LL, Wu LL, Li X, et al. Labeling and single-particle-tracking-based entry mechanism study of vaccinia virus from the Tiantan strain[J]. Anal Chem, 2018, 90(5): 3452-3459.
doi: 10.1021/acs.analchem.7b05183 pmid: 29392930 |
| [38] |
Zhang S, Chan KR, Tan HC, et al. Dengue virus growth, purification, and fluorescent labeling[J]. Methods Mol Biol, 2014, 1138: 3-14.
doi: 10.1007/978-1-4939-0348-1_1 pmid: 24696327 |
| [39] | Jha NK, Latinovic O, Martin E, et al. Imaging single retrovirus entry through alternative receptor isoforms and intermediates of virus-endosome fusion[J]. PLoS Pathog, 2011, 7(1): e1001260. |
| [40] | De Burghgraeve T, Kaptein SJF, Ayala-Nunez NV, et al. An analogue of the antibiotic teicoplanin prevents flavivirus entry in vitro[J]. PLoS One, 2012, 7(5): e37244. |
| [41] |
Hoornweg TE, Ayala Nuñez NV, et al. Dynamics of chikungunya virus cell entry unraveled by single-virus tracking in living cells[J]. J Virol, 2016, 90(9): 4745-4756.
doi: 10.1128/JVI.03184-15 pmid: 26912616 |
| [42] |
Floyd DL, Ragains JR, Skehel JJ, et al. Single-particle kinetics of influenza virus membrane fusion[J]. Proc Natl Acad Sci USA, 2008, 105(40): 15382-15387.
doi: 10.1073/pnas.0807771105 pmid: 18829437 |
| [43] |
Kremser L, Petsch M, Blaas D, et al. Labeling of capsid proteins and genomic RNA of human rhinovirus with two different fluorescent dyes for selective detection by capillary electrophoresis[J]. Anal Chem, 2004, 76(24): 7360-7365.
pmid: 15595880 |
| [44] |
Wen L, Lin Y, Zhang ZL, et al. Intracellular self-assembly based multi-labeling of key viral components: envelope, capsid and nucleic acids[J]. Biomaterials, 2016, 99: 24-33.
doi: 10.1016/j.biomaterials.2016.04.038 pmid: 27209260 |
| [45] |
Oscorbin IP, Belousova EA, Zakabunin AI, et al. Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification(qLAMP)[J]. BioTechniques, 2016, 61(1): 20-25.
doi: 10.2144/000114432 pmid: 27401670 |
| [46] |
Liu SL, Tian ZQ, Zhang ZL, et al. High-efficiency dual labeling of influenza virus for single-virus imaging[J]. Biomaterials, 2012, 33(31): 7828-7833.
doi: 10.1016/j.biomaterials.2012.07.026 URL |
| [47] |
Huang LL, Zhou P, Wang HZ, et al. A new stable and reliable method for labeling nucleic acids of fully replicative viruses[J]. Chem Commun, 2012, 48(18): 2424-2426.
doi: 10.1039/c2cc17069h URL |
| [48] |
Christensen N, Tilsner J, Bell K, et al. The 5’ cap of tobacco mosaic virus(TMV)is required for virion attachment to the actin/endoplasmic reticulum network during early infection[J]. Traffic, 2009, 10(5): 536-551.
doi: 10.1111/j.1600-0854.2009.00889.x pmid: 19220815 |
| [49] |
Xu HJ, Hao X, Wang SW, et al. Real-time imaging of rabies virus entry into living vero cells[J]. Sci Rep, 2015, 5: 11753.
doi: 10.1038/srep11753 pmid: 26148807 |
| [50] | Zhang S, Tan HC, Ooi EE. Visualizing dengue virus through Alexa fluor labeling[J]. J Vis Exp, 2011(53): e3168. |
| [51] |
Berlier JE, Rothe A, Buller G, et al. Quantitative comparison of long-wavelength Alexa fluor dyes to cy dyes: fluorescence of the dyes and their bioconjugates[J]. J Histochem Cytochem, 2003, 51(12): 1699-1712.
pmid: 14623938 |
| [52] |
Zhang SLX, Tan HC, Hanson BJ, et al. A simple method for Alexa Fluor dye labelling of dengue virus[J]. J Virol Methods, 2010, 167(2): 172-177.
doi: 10.1016/j.jviromet.2010.04.001 URL |
| [53] |
Fecek RJ, Busch R, Lin H, et al. Production of Alexa Fluor 488-labeled reovirus and characterization of target cell binding, competence, and immunogenicity of labeled virions[J]. J Immunol Methods, 2006, 314(1-2): 30-37.
doi: 10.1016/j.jim.2006.05.008 URL |
| [54] |
Gonçalves MS. Fluorescent labeling of biomolecules with organic probes[J]. Chem Rev, 2009, 109(1): 190-212.
doi: 10.1021/cr0783840 pmid: 19105748 |
| [55] |
王飞, 杨海涛, 王泽方. 红色荧光蛋白的研究进展[J]. 生物技术通报, 2017, 33(9): 32-47.
doi: 10.13560/j.cnki.biotech.bull.1985.2017-0401 |
|
Wang F, Yang HT, Wang ZF. Research progress on red fluorescent protein[J]. Biotechnol Bull, 2017, 33(9): 32-47.
doi: 10.13560/j.cnki.biotech.bull.1985.2017-0401 |
|
| [56] |
Stepanenko OV, Verkhusha VV, Kuznetsova IM, et al. Fluorescent proteins as biomarkers and biosensors: throwing color lights on molecular and cellular processes[J]. Curr Protein Pept Sci, 2008, 9(4): 338-369.
doi: 10.2174/138920308785132668 URL |
| [57] |
Lukyanov KA. Fluorescent proteins for a brighter science[J]. Biochem Biophys Res Commun, 2022, 633: 29-32.
doi: 10.1016/j.bbrc.2022.08.089 URL |
| [58] |
Mishin AS, Belousov VV, Solntsev KM, et al. Novel uses of fluorescent proteins[J]. Curr Opin Chem Biol, 2015, 27: 1-9.
doi: 10.1016/j.cbpa.2015.05.002 pmid: 26022943 |
| [59] | Bergeman MH, Hernandez MQ, Diefenderfer J, et al. Live-cell fluorescence microscopy of hsv-1 cellular egress by exocytosis[J]. bioRxiv, 2023. |
| [60] |
Wang HL, Wang Y, Giesman-Cookmeyer D, et al. Mutations in viral movement protein alter systemic infection and identify an intercellular barrier to entry into the phloem long-distance transport system[J]. Virology, 1998, 245(1): 75-89.
pmid: 9614869 |
| [61] |
Citovsky V. Tobacco mosaic virus: a pioneer of cell-to-cell movement[J]. Philos Trans R Soc Lond B Biol Sci, 1999, 354(1383): 637-643.
doi: 10.1098/rstb.1999.0415 URL |
| [62] |
Dai ZJ, He RR, Bernards MA, et al. The cis-expression of the coat protein of turnip mosaic virus is essential for viral intercellular movement in plants[J]. Mol Plant Pathol, 2020, 21(9): 1194-1211.
doi: 10.1111/mpp.12973 pmid: 32686275 |
| [63] |
Matevz R, Florence F, Michel T, et al. Fluorescently tagged potato virus Y: a versatile tool for functional analysis of plant-virus interactions[J]. Mol Plant Microbe Interact, 2015, 28(7): 739-750.
doi: 10.1094/MPMI-07-14-0218-TA URL |
| [64] |
Chudakov DM, Matz MV, Lukyanov S, et al. Fluorescent proteins and their applications in imaging living cells and tissues[J]. Physiol Rev, 2010, 90(3): 1103-1163.
doi: 10.1152/physrev.00038.2009 pmid: 20664080 |
| [65] |
Nienhaus GU, Wiedenmann J. Structure, dynamics and optical properties of fluorescent proteins: perspectives for marker development[J]. Chemphyschem, 2009, 10(9-10): 1369-1379.
doi: 10.1002/cphc.200800839 pmid: 19229892 |
| [66] |
Wang ZG, Liu SL, Pang DW. Quantum dots: a promising fluorescent label for probing virus trafficking[J]. Acc Chem Res, 2021, 54(14): 2991-3002.
doi: 10.1021/acs.accounts.1c00276 URL |
| [67] |
Liu HY, Wang ZG, Liu SL, et al. Single-virus tracking with quantum dots in live cells[J]. Nat Protoc, 2023, 18(2): 458-489.
doi: 10.1038/s41596-022-00775-7 |
| [68] |
Xu FZ, Qing TP, Qing ZH. DNA-coded metal nano-fluorophores: preparation, properties and applications in biosensing and bioimaging[J]. Nano Today, 2021, 36: 101021.
doi: 10.1016/j.nantod.2020.101021 URL |
| [69] | Wu QM, Liu SL, Chen G, et al. Uncovering the Rab5-independent autophagic trafficking of influenza A virus by quantum-dot-based single-virus tracking[J]. Small, 2018, 14(12): e1702841. |
| [70] |
Qin C, Li W, Li Q, et al. Real-time dissection of dynamic uncoating of individual influenza viruses[J]. Proc Natl Acad Sci USA, 2019, 116(7): 2577-2582.
doi: 10.1073/pnas.1812632116 pmid: 30626642 |
| [71] |
Yang YB, Tang YD, Hu Y, et al. Single virus tracking with quantum dots packaged into enveloped viruses using CRISPR[J]. Nano Lett, 2020, 20(2): 1417-1427.
doi: 10.1021/acs.nanolett.9b05103 URL |
| [72] |
Bergamaschi G, Metrangolo P, Dichiarante V. Photoluminescent nanocluster-based probes for bioimaging applications[J]. Photochem Photobiol Sci, 2022, 21(5): 787-801.
doi: 10.1007/s43630-021-00153-4 |
| [73] |
Zhang LB, Wang EK. Metal nanoclusters: new fluorescent probes for sensors and bioimaging[J]. Nano Today, 2014, 9(1): 132-157.
doi: 10.1016/j.nantod.2014.02.010 URL |
| [74] |
Zhang S, Zhang XY, Su ZQ. Biomolecule conjugated metal nanoclusters: bio-inspiration strategies, targeted therapeutics, and diagnostics[J]. J Mater Chem B, 2020, 8(19): 4176-4194.
doi: 10.1039/c9tb02936b pmid: 32313903 |
| [75] |
Tang J, Shi HH, Ma GY, et al. Ultrasmall Au and Ag nanoclusters for biomedical applications: a review[J]. Front Bioeng Biotechnol, 2020, 8: 1019.
doi: 10.3389/fbioe.2020.01019 URL |
| [76] | Kang J, Tahir A, Wang HJ, et al. Applications of nanotechnology in virus detection, tracking, and infection mechanisms[J]. Wiley Interdiscip Rev Nanomed Nanobiotechnol, 2021, 13(4): e1700. |
| [77] |
Marjomäki V, Lahtinen T, Martikainen M, et al. Site-specific targeting of enterovirus capsid by functionalized monodisperse gold nanoclusters[J]. Proc Natl Acad Sci USA, 2014, 111(4): 1277-1281.
doi: 10.1073/pnas.1310973111 pmid: 24474748 |
| [78] |
Wan XY, Zheng LL, Gao PF, et al. Real-time light scattering tracking of gold nanoparticles- bioconjugated respiratory syncytial virus infecting HEp-2 cells[J]. Sci Rep, 2014, 4: 4529.
doi: 10.1038/srep04529 |
| [79] |
Si P, Razmi N, Nur O, et al. Gold nanomaterials for optical biosensing and bioimaging[J]. Nanoscale Adv, 2021, 3(10): 2679-2698.
doi: 10.1039/d0na00961j pmid: 36134176 |
| [80] |
Chen PC, Mwakwari SC, Oyelere AK. Gold nanoparticles: from nanomedicine to nanosensing[J]. Nanotechnol Sci Appl, 2008, 1: 45-65.
pmid: 24198460 |
| [81] | 秦启伟. 单病毒示踪技术在病毒侵染机制及水生动物病毒研究中的应用进展[J]. 大连海洋大学学报, 2021, 36(6): 899-909. |
| Qin QW. Application of single-virus tracking technology on study of virus infection in live cells: a review[J]. J Dalian Ocean Univ, 2021, 36(6): 899-909. | |
| [82] |
Liu D, Pan L, Zhai HJ, et al. Virus tracking technologies and their applications in viral life cycle: research advances and future perspectives[J]. Front Immunol, 2023, 14: 1204730.
doi: 10.3389/fimmu.2023.1204730 URL |
| [83] | 李宇哲, 李阳阳, 沈熹涓, 等. SARS-CoV-2入胞活细胞示踪平台的建立[J]. 病毒学报, 2023, 39(3): 644-653. |
| Li YZ, Li YY, Shen XJ, et al. Establishment of A live-cell tracking platform for SARS-CoV-2 entry[J]. Chin J Virol, 2023, 39(3): 644-653. | |
| [84] |
Ivancová I, Leone DL, Hocek M. Reactive modifications of DNA nucleobases for labelling, bioconjugations, and cross-linking[J]. Curr Opin Chem Biol, 2019, 52: 136-144.
doi: S1367-5931(19)30067-5 pmid: 31415984 |
| [85] |
Kozoriz K, Shkel O, Hong KT, et al. Multifunctional photo-cross-linking probes: from target protein searching to imaging applications[J]. Acc Chem Res, 2023, 56(1): 25-36.
doi: 10.1021/acs.accounts.2c00505 URL |
| [86] |
Ganz D, Harijan D, Wagenknecht HA. Labelling of DNA and RNA in the cellular environment by means of bioorthogonal cycloaddition chemistry[J]. RSC Chem Biol, 2020, 1(3): 86-97.
doi: 10.1039/D0CB00047G URL |
| [87] |
Hong ZY, Lv C, Liu AN, et al. Clicking hydrazine and aldehyde: the way to labeling of viruses with quantum dots[J]. ACS Nano, 2015, 9(12): 11750-11760.
doi: 10.1021/acsnano.5b03256 URL |
| [88] |
Joo KI, Fang Y, Liu YR, et al. Enhanced real-time monitoring of adeno-associated virus trafficking by virus-quantum dot conjugates[J]. ACS Nano, 2011, 5(5): 3523-3535.
doi: 10.1021/nn102651p URL |
| [89] |
Jewett JC, Bertozzi CR. Cu-free click cycloaddition reactions in chemical biology[J]. Chem Soc Rev, 2010, 39(4): 1272-1279.
pmid: 20349533 |
| [90] |
Sletten EM, Bertozzi CR. From mechanism to mouse: a tale of two bioorthogonal reactions[J]. Acc Chem Res, 2011, 44(9): 666-676.
doi: 10.1021/ar200148z URL |
| [91] | Neumann S, Biewend M, Rana S, et al. The CuAAC: principles, homogeneous and heterogeneous catalysts, and novel developments and applications[J]. Macromol Rapid Commun, 2020, 41(1): e1900359. |
| [92] | Yang MY, Yang Y, Chen PR. Transition-metal-catalyzed bioorthogonal cycloaddition reactions[J]. Top Curr Chem(Cham), 2016, 374(1): 2. |
| [93] |
Anderson CT, Wallace IS, Somerville CR. Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls[J]. Proc Natl Acad Sci U S A, 2012, 109(4):1329-1334.
doi: 10.1073/pnas.1120429109 URL |
| [94] | 何宏星, 廖乃顺. 基于生物正交点击化学修饰吲哚菁绿用于脂肪间充质干细胞活体示踪的实验研究[J]. 中国实验动物学报, 2023, 31(8): 993-998. |
| He HX, Liao NS. In vivo tracking of adipose tissue-derived mesenchymal stem cells labeled with indocyanine green using bio-orthogonal click chemistry[J]. Acta Lab Animalis Sci Sin, 2023, 31(8): 993-998. | |
| [95] |
Ropitaux M, Hays Q, Baron A, et al. Dynamic imaging of cell wall polysaccharides by metabolic click-mediated labeling of pectins in living elongating cells[J]. Plant J, 2022, 110(3): 916-924.
doi: 10.1111/tpj.v110.3 URL |
| [96] |
Hao J, Huang LL, Zhang R, et al. A mild and reliable method to label enveloped virus with quantum dots by copper-free click chemistry[J]. Anal Chem, 2012, 84(19): 8364-8370.
doi: 10.1021/ac301918t pmid: 22946933 |
| [97] |
Zheng JD, Yue R, Yang RH, et al. Visualization of zika virus infection via a light-initiated bio-orthogonal cycloaddition labeling strategy[J]. Front Bioeng Biotechnol, 2022, 10: 940511.
doi: 10.3389/fbioe.2022.940511 URL |
| [98] |
Liu SL, Zhang LJ, Wang ZG, et al. Globally visualizing the microtubule-dependent transport behaviors of influenza virus in live cells[J]. Anal Chem, 2014, 86(8): 3902-3908.
doi: 10.1021/ac500640u URL |
| [99] |
Sun EZ, Liu AN, Zhang ZL, et al. Real-time dissection of distinct dynamin-dependent endocytic routes of influenza A virus by quantum dot-based single-virus tracking[J]. ACS Nano, 2017, 11(5): 4395-4406.
doi: 10.1021/acsnano.6b07853 URL |
| [100] | Zheng LL, Yang XX, Liu Y, et al. In situ labelling chemistry of respiratory syncytial viruses by employing the biotinylated host-cell membrane protein for tracking the early stage of virus entry[J]. Chem Commun(Camb), 2014, 50(99):15776-15779. |
| [101] |
Liu JF, Su MH, Chen X, et al. Lipid-mediated biosynthetic labeling strategy for in vivo dynamic tracing of avian influenza virus infection[J]. J Biomater Appl, 2022, 36(9): 1689-1699.
doi: 10.1177/08853282211063298 URL |
| [102] | Wen L, Zheng ZH, Liu AA, et al. Tracking single baculovirus retrograde transportation in host cell via quantum dot-labeling of virus internal component[J]. J Nanobiotechnology, 2017, 15(1): 37. |
| [103] |
Los GV, Encell LP, McDougall MG, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis[J]. ACS Chem Biol, 2008, 3(6): 373-382.
doi: 10.1021/cb800025k pmid: 18533659 |
| [104] |
Gong YK, Pan LF. Recent advances in bioorthogonal reactions for site-specific protein labeling and engineering[J]. Tetrahedron Lett, 2015, 56(17): 2123-2132.
doi: 10.1016/j.tetlet.2015.03.065 URL |
| [105] | Shah R, Lan SY, Puray-Chavez MN, et al. Single-cell multiplexed fluorescence imaging to visualize viral nucleic acids and proteins and monitor HIV, HTLV, HBV, HCV, zika virus, and influenza infection[J]. J Vis Exp, 2020(164): 10.3791/61843. |
| [106] |
Liu AA, Zhang ZF, Sun EZ, et al. Simultaneous visualization of parental and progeny viruses by a capsid-specific HaloTag labeling strategy[J]. ACS Nano, 2016, 10(1): 1147-1155.
doi: 10.1021/acsnano.5b06438 URL |
| [107] |
Ma YX, Mao GB, Huang WR, et al. Quantum dot nanobeacons for single RNA labeling and imaging[J]. J Am Chem Soc, 2019, 141(34): 13454-13458.
doi: 10.1021/jacs.9b04659 pmid: 31339040 |
| [108] |
Chiu YF, Huang YW, Chen CY, et al. Visualizing influenza A virus vRNA replication[J]. Front Microbiol, 2022, 13: 812711.
doi: 10.3389/fmicb.2022.812711 URL |
| [109] |
Zhang Y, Ke XL, Zheng ZH, et al. Encapsulating quantum dots into enveloped virus in living cells for tracking virus infection[J]. ACS Nano, 2013, 7(5): 3896-3904.
doi: 10.1021/nn305189n pmid: 23560365 |
| [110] | Elliott AD. Confocal microscopy: principles and modern practices[J]. Curr Protoc Cytom, 2020, 92(1): e68. |
| [111] |
Manivanh R, Lakdawala SS, Jones JE. Three-dimensional simultaneous imaging of nucleic acids and proteins during influenza virus infection in single cells using confocal microscopy[J]. Methods Mol Biol, 2022, 2440: 41-56.
doi: 10.1007/978-1-0716-2051-9_2 pmid: 35218531 |
| [112] | Pina MAS, Gómez-Aix C, Méndez-López E, et al. Imaging techniques to study plant virus replication and vertical transmission[J]. Viruses, 2021, 13(3): 358. |
| [113] | Fish KN. Total internal reflection fluorescence(TIRF)microscopy[J]. Curr Protoc Cytom, 2009, 50(1): 12.18.1-12.18.13. |
| [114] | Martín-Cófreces NB, Torralba D, Lozano-Prieto M, et al. TIRF microscopy as a tool to determine exosome composition[J]. Methods Mol Biol, 2021, 2346: 91-104. |
| [115] |
Bally M, Block S, Höök F, et al. Physicochemical tools for studying virus interactions with targeted cell membranes in a molecular and spatiotemporally resolved context[J]. Anal Bioanal Chem, 2021, 413(29): 7157-7178.
doi: 10.1007/s00216-021-03510-5 pmid: 34490501 |
| [116] | Bernacchi S. Visualization of retroviral gag-genomic RNA cellular interactions leading to genome encapsidation and viral assembly: an overview[J]. Viruses, 2022, 14(2): 324. |
| [117] | van der Borg G, Braddock S, Blijleven JS, et al. Single-particle fusion of influenza viruses reveals complex interactions with target membranes[J]. J Phys Condens Matter, 2018, 30(20): 204005. |
| [118] | Rust MJ, Bates M, Zhuang XW. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy(STORM)[J]. Nat Methods, 2006, 3(10): 793-795. |
| [119] |
Huang B, Wang WQ, Bates M, et al. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy[J]. Science, 2008, 319(5864): 810-813.
doi: 10.1126/science.1153529 pmid: 18174397 |
| [120] | Xu JQ, Ma HQ, Liu Y. Stochastic optical reconstruction microscopy(STORM)[J]. CP Cytometry, 2017, 81(1): 12.46.1-12.46.27. |
| [121] | Zhang XN, Yue L, Zhang ZQ, et al. Establishment of a fluorescent in situ hybridization assay for imaging hepatitis B virus nucleic acids in cell culture models[J]. Emerg Microbes Infect, 2017, 6(11): e98. |
| [122] |
Feng H, Wang XB, Xu ZW, et al. Super-resolution fluorescence microscopy for single cell imaging[J]. Adv Exp Med Biol, 2018, 1068: 59-71.
doi: 10.1007/978-981-13-0502-3_6 pmid: 29943296 |
| [123] |
Sakin V, Paci G, Lemke EA, et al. Labeling of virus components for advanced, quantitative imaging analyses[J]. FEBS Lett, 2016, 590(13): 1896-1914.
doi: 10.1002/1873-3468.12131 pmid: 26987299 |
| [124] | Rust MJ, Lakadamyali M, Brandenburg B, et al. Single-virus tracking in live cells[J]. Cold Spring Harb Protoc, 2011, 2011(9): pdb.top065623. |
| [125] |
Schermelleh L, Ferrand A, Huser T, et al. Super-resolution microscopy demystified[J]. Nat Cell Biol, 2019, 21(1): 72-84.
doi: 10.1038/s41556-018-0251-8 pmid: 30602772 |
| [126] |
Huang LL, Xie HY. Progress on the labeling and single-particle tracking technologies of viruses[J]. Analyst, 2014, 139(13): 3336-3346.
doi: 10.1039/C4AN00038B URL |
| [127] |
Qu XH, Wu D, Mets L, et al. Nanometer-localized multiple single-molecule fluorescence microscopy[J]. Proc Natl Acad Sci USA, 2004, 101(31): 11298-11303.
pmid: 15277661 |
| [128] |
Patwardhan A. Subpixel position measurement using 1D, 2D and 3D centroid algorithms with emphasis on applications in confocal microscopy[J]. J Microsc, 2003, 186(3): 246-257.
doi: 10.1046/j.1365-2818.1997.1970761.x URL |
| [129] |
Ruthardt N, Lamb DC, Bräuchle C. Single-particle tracking as a quantitative microscopy-based approach to unravel cell entry mechanisms of viruses and pharmaceutical nanoparticles[J]. Mol Ther, 2011, 19(7): 1199-1211.
doi: 10.1038/mt.2011.102 pmid: 21654634 |
| [130] |
Small A, Stahlheber S. Fluorophore localization algorithms for super-resolution microscopy[J]. Nat Methods, 2014, 11(3): 267-279.
doi: 10.1038/nmeth.2844 pmid: 24577277 |
| [131] |
Sage D, Neumann FR, Hediger F, et al. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics[J]. IEEE Trans Image Process, 2005, 14(9): 1372-1383.
doi: 10.1109/TIP.2005.852787 URL |
| [132] |
Feng LQ, Xu YK, Yang Y, et al. Multiple dense particle tracking in fluorescence microscopy images based on multidimensional assignment[J]. J Struct Biol, 2011, 173(2): 219-228.
doi: 10.1016/j.jsb.2010.11.001 pmid: 21073957 |
| [133] |
Smal I, Meijering E. Quantitative comparison of multiframe data association techniques for particle tracking in time-lapse fluorescence microscopy[J]. Med Image Anal, 2015, 24(1): 163-189.
doi: S1361-8415(15)00093-6 pmid: 26176413 |
| [134] |
Jaqaman K, Loerke D, Mettlen M, et al. Robust single-particle tracking in live-cell time-lapse sequences[J]. Nat Methods, 2008, 5(8): 695-702.
doi: 10.1038/nmeth.1237 pmid: 18641657 |
| [135] |
Smal I, Meijering E, Draegestein K, et al. Multiple object tracking in molecular bioimaging by Rao-Blackwellized marginal particle filtering[J]. Med Image Anal, 2008, 12(6): 764-777.
doi: 10.1016/j.media.2008.03.004 pmid: 18457985 |
| [136] |
Chenouard N, Bloch I, Olivo-Marin JC. Multiple hypothesis tracking for cluttered biological image sequences[J]. IEEE Trans Pattern Anal Mach Intell, 2013, 35(11): 2736-3750.
doi: 10.1109/TPAMI.2013.97 URL |
| [1] | ZHANG Xiao-ni, WENG Yi-chun, FAN Yi-hao, WANG Xiao-juan, ZHAO Jia-yu, ZHANG Yun-long. Mito-OS-Timer:A Targeted Fluorescent Stopwatch for Monitoring Mitochondrial Oxidative Stress [J]. Biotechnology Bulletin, 2022, 38(10): 97-105. |
| [2] | WANG Huan-yu, CHANG Hao-wan, ZHANG Chong-qi, JIN Wei-lin, WEI Fang. Comparison of 5 Methods of Evaluating the Expressions of Chimeric Antigen Receptors [J]. Biotechnology Bulletin, 2021, 37(12): 265-273. |
| [3] | ZHOU Jia-sheng, WANG Peng, ZHOU Huang-mei, XU Jin-ming, ZHANG San-jun. Progress in Fluorescence Lifetime Imaging of Circularly-permuted Fluorescent Protein Biosensors [J]. Biotechnology Bulletin, 2018, 34(10): 71-80. |
| [4] | WANG Fei,YANG Hai-tao,WANG Ze-fang. Research Progress on Red Fluorescent Protein [J]. Biotechnology Bulletin, 2017, 33(9): 32-47. |
| [5] | CHEN Nan, YU Fei, HE Yan-liu, BU Ning. Labeling and Tracing of Green Fluorescent Protein in Fungal Endophyte with Growth-promoting Activity to Rice Seedlings [J]. Biotechnology Bulletin, 2017, 33(3): 100-105. |
| [6] | LIU Xiao-yu, MA Yu-chao. Green Fluorescent Protein Marker of Biocontrol Streptomyces SSD49 and Its Colonization on the Populus tomentosa Somaclone [J]. Biotechnology Bulletin, 2016, 32(9): 197-202. |
| [7] | WANG Hai-tao,JIN Bo,LI Shu-wei. Eukaryotic Expression of MSTN Gene from Hetian Sheep,Carla Kul Sheep,and Duolang Sheep in the Southern Xinjiang [J]. Biotechnology Bulletin, 2016, 32(5): 82-90. |
| [8] | Liang Junting, Li Luzhi, Chen Shaopeng, Jiao Zhen. Study on Sites of the Tolerate Peptide Insertion in the Fluorescent Protein of mCherry [J]. Biotechnology Bulletin, 2013, 0(5): 144-148. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||