








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (5): 38-47.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1135
Previous Articles Next Articles
XIAO Yi-meng( ), YANG Wen, CHENG Yi-yi, LUO Gang(
), YANG Wen, CHENG Yi-yi, LUO Gang( )
)
Received:2023-12-01
Online:2024-05-26
Published:2024-03-29
Contact:
LUO Gang
E-mail:xymxiaoyimeng@163.com;luo_gang@just.edu.cn
XIAO Yi-meng, YANG Wen, CHENG Yi-yi, LUO Gang. CRISPR-Cas9 Gene Editing Technology and Its Research Progress in Poultry[J]. Biotechnology Bulletin, 2024, 40(5): 38-47.

Fig. 1 Schematic diagram of the CRISPR-Cas9/dCas9 system cutting double-stranded DNA A: Schematic diagram of CRISPR-Cas9 cutting target DNA. Cas9: CRISPR-associated protein 9; RuvC and HNH: structural domains in the Cas9 protein that cleave nucleic acids; PAM sequence: protospacer adjacent motif; sgRNA: single-guide RNA, composed of crRNA(CRISPR RNA)and tracrRNA(trans-activating crRNA). B and C: Schematic representation of CRISPR-nCas9 cleaving target DNA. nCas9: Cas9 nickase; HNH(H840A): Cas9 mutant with inactivated HNH domain; RuvC(D10A): Cas9 mutant with inactivated RuvC domain. D: Schematic diagram of CRISPR-dCas9 cutting target DNA. HNH(H840A)and RuvC(D10A): Cas9 mutants with inactivated HNH and RuvC structural domains
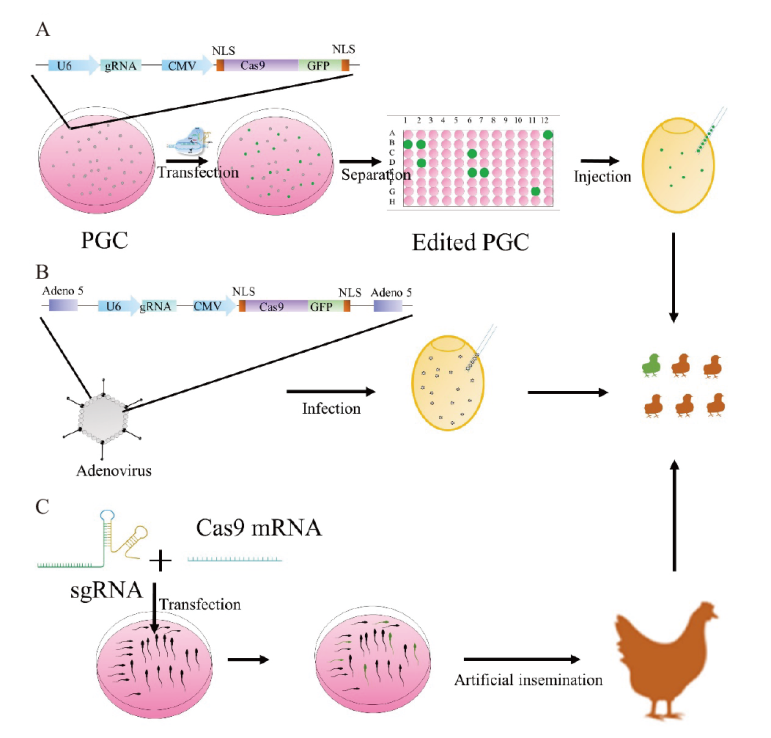
Fig. 2 Delivery of the CRISPR-Cas9 system in avian species A: Schematic diagram of PGC-mediated CRISPR-Cas9 gene editing. U6: gRNA promoter; gRNA: guide RNA; CMV: The promoter of Cas9-encoded genes; GFP: green fluorescent protein; NLS: nuclear localization sequences; PGC: primordial germ cell. B: Schematic diagram of adenovirus-mediated CRISPR-Cas9 gene editing. C: Schematic representation of STAGE-mediated CRISPR-Cas9. STAGE: sperm transfection-assisted gene editing; sgRNA: single guide RNA; Cas 9 mRNA: messenger RNA of Cas 9-encoding genes
| [1] |
Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and Archaea: versatile small RNAs for adaptive defense and regulation[J]. Annu Rev Genet, 2011, 45: 273-297.
doi: 10.1146/annurev-genet-110410-132430 pmid: 22060043 |
| [2] |
Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823.
doi: 10.1126/science.1231143 pmid: 23287718 |
| [3] |
Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information[J]. Virology, 2012, 434(2): 202-209.
doi: 10.1016/j.virol.2012.10.003 pmid: 23123013 |
| [4] | Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096. |
| [5] | Jinek M, East A, Cheng A, et al. RNA-programmed genome editing in human cells[J]. eLife, 2013, 2: e00471. |
| [6] |
Shmakov S, Smargon A, Scott D, et al. Diversity and evolution of class 2 CRISPR-Cas systems[J]. Nat Rev Microbiol, 2017, 15(3): 169-182.
doi: 10.1038/nrmicro.2016.184 pmid: 28111461 |
| [7] | Ding WT, Zhang Y, Shi SB. Development and application of CRISPR/cas in microbial biotechnology[J]. Front Bioeng Biotechnol, 2020, 8: 711. |
| [8] |
Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression[J]. Cell, 2021, 184(3): 844.
doi: 10.1016/j.cell.2021.01.019 pmid: 33545038 |
| [9] |
Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nat Biotechnol, 2020, 38(7): 824-844.
doi: 10.1038/s41587-020-0561-9 pmid: 32572269 |
| [10] |
Collias D, Beisel CL. CRISPR technologies and the search for the PAM-free nuclease[J]. Nat Commun, 2021, 12(1): 555.
doi: 10.1038/s41467-020-20633-y pmid: 33483498 |
| [11] | Kleinstiver BP, Prew MS, Tsai SQ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities[J]. Nature, 2015, 523(7561): 481-485. |
| [12] | Chatterjee P, Jakimo N, Jacobson JM. Minimal PAM specificity of a highly similar SpCas9 ortholog[J]. Sci Adv, 2018, 4(10): eaau0766. |
| [13] |
Chatterjee P, Lee J, Nip L, et al. A Cas9 with PAM recognition for adenine dinucleotides[J]. Nat Commun, 2020, 11(1): 2474.
doi: 10.1038/s41467-020-16117-8 pmid: 32424114 |
| [14] |
Miller SM, Wang TN, Randolph PB, et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs[J]. Nat Biotechnol, 2020, 38(4): 471-481.
doi: 10.1038/s41587-020-0412-8 pmid: 32042170 |
| [15] | Christie KA, Guo JA, Silverstein RA, et al. Precise DNA cleavage using CRISPR-SpRYgests[J]. Nat Biotechnol, 2023, 41(3): 409-416. |
| [16] |
Walton RT, Christie KA, Whittaker MN, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants[J]. Science, 2020, 368(6488): 290-296.
doi: 10.1126/science.aba8853 pmid: 32217751 |
| [17] | Saito M, Xu PY, Faure G, et al. Fanzor is a eukaryotic programmable RNA-guided endonuclease[J]. Nature, 2023, 620(7974): 660-668. |
| [18] |
Chen BH, Gilbert LA, Cimini BA, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system[J]. Cell, 2013, 155(7): 1479-1491.
doi: 10.1016/j.cell.2013.12.001 pmid: 24360272 |
| [19] |
Kudo T, Sutou S. Usage of putative chicken U6 promoters for vector-based RNA interference[J]. J Reprod Dev, 2005, 51(3): 411-417.
pmid: 15812142 |
| [20] |
Lee J, Ma JS, Lee K. Direct delivery of adenoviral CRISPR/Cas9 vector into the blastoderm for generation of targeted gene knockout in quail[J]. Proc Natl Acad Sci USA, 2019, 116(27): 13288-13292.
doi: 10.1073/pnas.1903230116 pmid: 31209054 |
| [21] | Lee J, Kim DH, Karolak MC, et al. Generation of genome-edited chicken and duck lines by adenovirus-mediated in vivo genome editing[J]. Proc Natl Acad Sci USA, 2022, 119(45): e2214344119. |
| [22] |
Gassler T, Heistinger L, Mattanovich D, et al. CRISPR/Cas9-mediated homology-directed genome editing in Pichia pastoris[J]. Methods Mol Biol, 2019, 1923: 211-225.
doi: 10.1007/978-1-4939-9024-5_9 pmid: 30737742 |
| [23] | Gao YB, Zhao YD. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing[J]. J Integr Plant Biol, 2014, 56(4): 343-349. |
| [24] |
Wang JQ, Li X, Zhao YH, et al. Generation of cell-type-specific gene mutations by expressing the sgRNA of the CRISPR system from the RNA polymerase II promoters[J]. Protein Cell, 2015, 6(9): 689-692.
doi: 10.1007/s13238-015-0169-x pmid: 26050089 |
| [25] |
Mashiko D, Fujihara Y, Satouh Y, et al. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA[J]. Sci Rep, 2013, 3: 3355.
doi: 10.1038/srep03355 pmid: 24284873 |
| [26] |
Pei WH, Burgess SM. Microinjection in zebrafish for genome editing and functional studies[J]. Methods Mol Biol, 2019, 1874: 459-474.
doi: 10.1007/978-1-4939-8831-0_26 pmid: 30353530 |
| [27] | Han JY, Park YH. Primordial germ cell-mediated transgenesis and genome editing in birds[J]. J Anim Sci Biotechnol, 2018, 9: 19. |
| [28] |
Schusser B, Collarini EJ, Yi H, et al. Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells[J]. Proc Natl Acad Sci USA, 2013, 110(50): 20170-20175.
doi: 10.1073/pnas.1317106110 pmid: 24282302 |
| [29] |
Oishi I, Yoshii K, Miyahara D, et al. Targeted mutagenesis in chicken using CRISPR/Cas9 system[J]. Sci Rep, 2016, 6: 23980.
doi: 10.1038/srep23980 pmid: 27050479 |
| [30] |
Lee HJ, Yoon JW, Jung KM, et al. Targeted gene insertion into Z chromosome of chicken primordial germ cells for avian sexing model development[J]. FASEB J, 2019, 33(7): 8519-8529.
doi: 10.1096/fj.201802671R pmid: 30951374 |
| [31] |
Han JY, Lee BR. Isolation and characterization of chicken primordial germ cells and their application in transgenesis[J]. Methods Mol Biol, 2017, 1650: 229-242.
doi: 10.1007/978-1-4939-7216-6_15 pmid: 28809025 |
| [32] | Dimitrov L, Pedersen D, Ching KH, et al. Germline gene editing in chickens by efficient CRISPR-mediated homologous recombination in primordial germ cells[J]. PLoS One, 2016, 11(4): e0154303. |
| [33] | 孙娟娟. 利用CRISPR/Cas9技术表达鸡卵清融合蛋白的初步研究[D]. 南宁: 广西大学, 2019. |
| Sun JJ. Expression of ovalbumin fusion protein in chicken using CRISPR/Cas9 technology[D]. Nanning: Guangxi University, 2019. | |
| [34] | 彭腊如. 应用CRISPR/Cas9技术敲除鸡GDF8基因的研究[D]. 南宁: 广西大学, 2020. |
| Peng LR. Knocking out chicken GDF8 gene using CRISPR/Cas9 technology[D]. Nanning: Guangxi University, 2020. | |
| [35] | Tyack SG, Jenkins KA, O'Neil TE, et al. A new method for producing transgenic birds via direct in vivo transfection of primordial germ cells[J]. Transgenic Res, 2013, 22(6): 1257-1264. |
| [36] | Challagulla A, Jenkins KA, O'Neil TE, et al. Germline engineering of the chicken genome using CRISPR/Cas9 by in vivo transfection of PGCs[J]. Anim Biotechnol, 2023, 34(4): 775-784. |
| [37] | Kang KS, Lee HC, Kim HJ, et al. Spatial and temporal action of chicken primordial germ cells during initial migration[J]. Reproduction, 2015, 149(2): 179-187. |
| [38] | Ahn J, Lee J, Park JY, et al. Targeted genome editing in a quail cell line using a customized CRISPR/Cas9 system[J]. Poult Sci, 2017, 96(5): 1445-1450. |
| [39] | Lee J, Kim DH, Lee K. Muscle hyperplasia in Japanese quail by single amino acid deletion in MSTN propeptide[J]. Int J Mol Sci, 2020, 21(4): 1504. |
| [40] | 胡豪. CRISPR/Cas9腺病毒直接注射法制备肌肉抑制素(MSTN)基因敲除鸡的研究初探[D]. 南宁: 广西大学, 2019. |
| Hu H. Preliminary study on the preparation of myostatin(MSTN)knockout chicken by direct injection of CRISPR/Cas9 adenovirus[D]. Nanning: Guangxi University, 2019. | |
| [41] | 梁传堉. 腺病毒介导SpCas9对鸡AMH基因高效敲除系统的构建[D]. 杨凌: 西北农林科技大学, 2019. |
| Liang CY. Construction of an adenovirus-mediated SpCas9 efficient knockout system for chicken AMH[D]. Yangling: Northwest A & F University, 2019. | |
| [42] | Lee J, Kim DH, Lee K. Research Note: injection of adenoviral CRISPR/Cas9 system targeting melanophilin gene into different sites of embryos induced regional feather color changes in posthatch quail[J]. Poult Sci, 2023, 102(11): 103087. |
| [43] |
Ball BA, Sabeur K, Allen WR. Liposome-mediated uptake of exogenous DNA by equine spermatozoa and applications in sperm-mediated gene transfer[J]. Equine Vet J, 2008, 40(1): 76-82.
pmid: 18083664 |
| [44] | Cooper CA, Challagulla A, Jenkins KA, et al. Generation of gene edited birds in one generation using sperm transfection assisted gene editing(STAGE)[J]. Transgenic Res, 2017, 26(3): 331-347. |
| [45] | Cooper CA, Doran TJ, Challagulla A, et al. Innovative approaches to genome editing in avian species[J]. J Anim Sci Biotechnol, 2018, 9: 15. |
| [46] | 代敏敏. 精子载体制备转基因鸡方法研究[D]. 保定: 河北农业大学, 2020. |
| Dai MM. Study on the method of preparing transgenic chicken with sperm carrier[D]. Baoding: Hebei Agricultural University, 2020. | |
| [47] |
Shin J, Bae DR, Latshaw JD, et al. Technical note: a gene delivery system in the embryonic cells of avian species using a human adenoviral vector[J]. J Anim Sci, 2009, 87(9): 2791-2795.
doi: 10.2527/jas.2009-1983 pmid: 19502509 |
| [48] |
Kim S, Kim D, Cho SW, et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins[J]. Genome Res, 2014, 24(6): 1012-1019.
doi: 10.1101/gr.171322.113 pmid: 24696461 |
| [49] |
Duan L, Ouyang K, Wang JH, et al. Exosomes as targeted delivery platform of CRISPR/Cas9 for therapeutic genome editing[J]. Chembiochem, 2021, 22(24): 3360-3368.
doi: 10.1002/cbic.202100359 pmid: 34418266 |
| [50] |
Gee P, Lung M, Okuzaki Y, et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping[J]. Nat Commun, 2020, 11(1): 1334.
doi: 10.1038/s41467-020-14957-y pmid: 32170079 |
| [51] | Kim D, Le QV, Wu YN, et al. Nanovesicle-mediated delivery systems for CRISPR/cas genome editing[J]. Pharmaceutics, 2020, 12(12): 1233. |
| [52] |
Chojnacka-Puchta L, Sawicka D. CRISPR/Cas9 gene editing in a chicken model: current approaches and applications[J]. J Appl Genet, 2020, 61(2): 221-229.
doi: 10.1007/s13353-020-00537-9 pmid: 31925767 |
| [53] | Zhu XX, Zhan QM, Wei YY, et al. CRISPR/Cas9-mediated MSTN disruption accelerates the growth of Chinese Bama pigs[J]. Reprod Domest Anim, 2020, 55(10): 1314-1327. |
| [54] | Kalds P, Crispo M, Li C, et al. Generation of double-muscled sheep and goats by CRISPR/Cas9-mediated knockout of the myostatin gene[J]. Methods Mol Biol, 2022, 2495: 295-323. |
| [55] |
Park TS, Park J, Lee JH, et al. Disruption of G0/G1 switch gene 2(G0S2)reduced abdominal fat deposition and altered fatty acid composition in chicken[J]. FASEB J, 2019, 33(1): 1188-1198.
doi: 10.1096/fj.201800784R pmid: 30085885 |
| [56] |
Zhang XD, Xie XT, Heckmann BL, et al. Targeted disruption of G0/G1 switch gene 2 enhances adipose lipolysis, alters hepatic energy balance, and alleviates high-fat diet-induced liver steatosis[J]. Diabetes, 2014, 63(3): 934-946.
doi: 10.2337/db13-1422 pmid: 24194501 |
| [57] |
Whitworth KM, Rowland R, Ewen CL, et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus[J]. Nat Biotechnol, 2016, 34(1): 20-22.
doi: 10.1038/nbt.3434 pmid: 26641533 |
| [58] | Koslová A, Kučerová D, Reinišová M, et al. Genetic resistance to avian leukosis viruses induced by CRISPR/Cas9 editing of specific receptor genes in chicken cells[J]. Viruses, 2018, 10(11): 605. |
| [59] |
Koslová A, Trefil P, Mucksová J, et al. Precise CRISPR/Cas9 editing of the NHE1 gene renders chickens resistant to the J subgroup of avian leukosis virus[J]. Proc Natl Acad Sci USA, 2020, 117(4): 2108-2112.
doi: 10.1073/pnas.1913827117 pmid: 31964810 |
| [60] | Long JS, Giotis ES, Moncorgé O, et al. Species difference in ANP32A underlies influenza A virus polymerase host restriction[J]. Nature, 2016, 529: 101-104. |
| [61] | Xie Q, Wang WK, Kan QQ, et al. FAdV-4 without Fiber-2 is a highly attenuated and protective vaccine candidate[J]. Microbiol Spectr, 2022, 10(1): e0143621. |
| [62] | Chang PX, Ameen F, Sealy JE, et al. Application of HDR-CRISPR/Cas9 and erythrocyte binding for rapid generation of recombinant Turkey herpesvirus-vectored avian influenza virus vaccines[J]. Vaccines, 2019, 7(4): 192. |
| [63] | Teng M, Zhou ZY, Yao YX, et al. A new strategy for efficient screening and identification of monoclonal antibodies against oncogenic avian herpesvirus utilizing CRISPR/Cas9-based gene-editing technology[J]. Viruses, 2022, 14(9): 2045. |
| [64] |
Pardee K, Green AA, Takahashi MK, et al. Rapid, low-cost detection of zika virus using programmable biomolecular components[J]. Cell, 2016, 165(5): 1255-1266.
doi: S0092-8674(16)30505-0 pmid: 27160350 |
| [65] | Srivastava S, Upadhyay DJ, Srivastava A. Next-generation molecular diagnostics development by CRISPR/cas tool: rapid detection and surveillance of viral disease outbreaks[J]. Front Mol Biosci, 2020, 7: 582499. |
| [66] |
Park JS, Choi HJ, Jung KM, et al. Production of recombinant human IgG1 Fc with beneficial N-glycosylation pattern for anti-inflammatory activity using genome-edited chickens[J]. Commun Biol, 2023, 6(1): 589.
doi: 10.1038/s42003-023-04937-5 pmid: 37264071 |
| [67] | Kim YM, Park JS, Choi HJ, et al. Efficient production of recombinant human adiponectin in egg white using genome edited chickens[J]. Front Nutr, 2023, 9: 1068558. |
| [68] |
Oishi I, Yoshii K, Miyahara D, et al. Efficient production of human interferon beta in the white of eggs from ovalbumin gene-targeted hens[J]. Sci Rep, 2018, 8(1): 10203.
doi: 10.1038/s41598-018-28438-2 pmid: 29976933 |
| [69] | Kim YM, Shim JH, Park JS, et al. Sequential verification of exogenous protein production in OVA gene-targeted chicken bioreactors[J]. Poult Sci, 2023, 102(1): 102247. |
| [70] | Zhang YN, Wang YJ, Zuo QS, et al. CRISPR/Cas9 mediated chicken Stra8 gene knockout and inhibition of male germ cell differentiation[J]. PLoS One, 2017, 12(2): e0172207. |
| [71] | Lee JH, Park JW, Kim SW, et al. C-X-C chemokine receptor type 4(CXCR4)is a key receptor for chicken primordial germ cell migration[J]. J Reprod Dev, 2017, 63(6): 555-562. |
| [72] | Lee KY, Choi HJ, Park KJ, et al. Development and characterization of a CRISPR/Cas9-mediated RAG1 knockout chicken model lacking mature B and T cells[J]. Front Immunol, 2022, 13: 892476. |
| [73] | Wang L, Xue Z, Wang JP, et al. Targeted knockout of Mx in the DF-1 chicken fibroblast cell line impairs immune response against Newcastle disease virus[J]. Poult Sci, 2023, 102(9): 102855. |
| [74] | Chapman B, Han JH, Lee HJ, et al. Targeted modulation of chicken genes in vitro using CRISPRa and CRISPRi toolkit[J]. Genes, 2023, 14(4): 906. |
| [75] | Williams RM, Senanayake U, Artibani M, et al. Genome and epigenome engineering CRISPR toolkit for in vivo modulation of cis-regulatory interactions and gene expression in the chicken embryo[J]. Development, 2018, 145(4): dev160333. |
| [1] | WANG Ling-ling, MA Ai-hong, SONG Qian-jin, WANG Xiao-long, CAO Xiao-zhen, LIU Zhi-feng, CHEN Ya-fei, LI Ji-dong. Establishment and Application of Dual RPA-LFD Detection Method for Capripox Virus and Orf Virus [J]. Biotechnology Bulletin, 2024, 40(5): 103-111. |
| [2] | WANG Xin, XU Yi-yi, XU Yang, XU Chen-wu. Research Progress in Genomic Selection Breeding Technology for Crops [J]. Biotechnology Bulletin, 2024, 40(3): 1-13. |
| [3] | HE Shi-yu, ZENG Zhong-da, LI Bo-yan. Application Progress of Spatially Resolved Metabolomics in Disease Diagnosis Research [J]. Biotechnology Bulletin, 2024, 40(1): 145-159. |
| [4] | ZHAN Yan, ZHOU Li-bin, JIN Wen-jie, DU Yan, YU Li-xia, QU Ying, MA Yong-gui, LIU Rui-yuan. Research Progress in Plant Leaf Color Mutation Induced by Radiation [J]. Biotechnology Bulletin, 2023, 39(8): 106-113. |
| [5] | ZHANG Dao-lei, GAN Yu-jun, LE Liang, PU Li. Epigenetic Regulation of Yield-related Traits in Maize and Epibreeding [J]. Biotechnology Bulletin, 2023, 39(8): 31-42. |
| [6] | LENG Yan, MA Xiao-wei, CHEN Guang, REN He, LI Xiang. High-yield Contests in Maize Facilitate the Vitalization of China’s Seed Industry [J]. Biotechnology Bulletin, 2023, 39(8): 4-10. |
| [7] | WANG Tian-yi, WANG Rong-huan, WANG Xia-qing, ZHANG Ru-yang, XU Rui-bin, JIAO Yan-yan, SUN Xuan, WANG Ji-dong, SONG Wei, ZHAO Jiu-ran. Research in Maize Dwarf Genes and Dwarf Breeding [J]. Biotechnology Bulletin, 2023, 39(8): 43-51. |
| [8] | LI Yu-ling, MAO Xin, ZHANG Yuan-shuai, DONG Yuan-fu, LIU Cui-lan, DUAN Chun-hua, MAO Xiu-hong. Applications and Perspectives of Radiation Mutagenesis in Woody Plant Breeding [J]. Biotechnology Bulletin, 2023, 39(6): 12-30. |
| [9] | ZHU Shao-xi, JIN Zhao-yang, GE Jian-rong, WANG Rui, WANG Feng-ge, LU Yun-cai. High-throughput Specific Detection Methods for Transgenic Maize Based on the KASP Platform [J]. Biotechnology Bulletin, 2023, 39(6): 133-140. |
| [10] | ZHANG Zu-lin, LIU Fang-fang, ZHOU Qing-niao, ZHAO Rui-qiang, HE Shu-jia, LIN Wen-zhen. Construction and Identification of Huh7 Hepatoma Cell Line with ACE2 Gene Knockout Based on CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(6): 181-188. |
| [11] | LEI Cai-rong, GUO Xiao-peng, CHAI Ran, ZHANG Miao-miao, REN Jun-le, LU Dong. Application of Omics Techniques in Incluced Breecling via Heavy Ion Beam Irradiating Microorganisms [J]. Biotechnology Bulletin, 2023, 39(5): 54-62. |
| [12] | XUE Jiao ZHU Qing-feng FENG Yan-zhao CHEN Pei LIU Wen-hua ZHANG Ai-xia LIU Qin-jian ZHANG Qi YU Yang. Advances in Upstream Open Reading Frame in Plant Genes [J]. Biotechnology Bulletin, 2023, 39(4): 157-165. |
| [13] | YANG Mao, LIN Yu-feng, DAI Yang-shuo, PAN Su-jun, PENG Wei-ye, YAN Ming-xiong, LI Wei, WANG Bing, DAI Liang-ying. OsDIS1 Negatively Regulates Rice Drought Tolerance Through Antioxidant Pathways [J]. Biotechnology Bulletin, 2023, 39(2): 88-95. |
| [14] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [15] | XIE Wei, LIU Chun-ming. Commercialization of Biological Breeding in China: Opportunities and Policy Issues [J]. Biotechnology Bulletin, 2023, 39(1): 16-20. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||