








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (4): 67-76.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1152
Previous Articles Next Articles
ZHANG Yi-heng1,2,3( ), LIU Jia-zheng4,5,6, WANG Xue-chen2,3, SUN Zheng-zhe7, XUE Ya-jun2,3, WANG Pei1, HAN Hua4,5,6, ZHENG Hong-wei8(
), LIU Jia-zheng4,5,6, WANG Xue-chen2,3, SUN Zheng-zhe7, XUE Ya-jun2,3, WANG Pei1, HAN Hua4,5,6, ZHENG Hong-wei8( ), LI Xiao-juan2,3(
), LI Xiao-juan2,3( )
)
Received:2023-12-08
Online:2024-04-26
Published:2024-04-30
Contact:
ZHENG Hong-wei, LI Xiao-juan
E-mail:zhangyh20@bjfu.edu.cn;hzheng@ms.xjb.ac.cn;lixj@bjfu.edu.cn
ZHANG Yi-heng, LIU Jia-zheng, WANG Xue-chen, SUN Zheng-zhe, XUE Ya-jun, WANG Pei, HAN Hua, ZHENG Hong-wei, LI Xiao-juan. Dynamic Changes of Arabidopsis Endoplasmic Reticulum Based on Enhanced Super-resolution Images[J]. Biotechnology Bulletin, 2024, 40(4): 67-76.

Fig. 1 Workflow for de-noising, enhancement, automatic identification and quantitative analysis based on the super-resolution imaging in the root zone of plant A: The structure and dynamics of ER(endoplasmic reticulum)in the root cells were studied in living material using Arabidopsis seedlings, with the region shown in the box as the elongation zone(Bar = 2 μm). B: ER images acquired by confocal microscopy. C: Imaging of ER using SIM. D: Imaging results of ER using SIM. E: Imaging results processed by utilizing the enhancement denoising framework. F: Segmentation of ER structural features on the acquired images using Swint-ResU-Net(green markers are bulk flow; red markers are fusiform body, darks blue markers are the tubules, and light blue markers are cisternaes). G: Statistics were done on the nine feature parameters quantified by the acquisition(the ratio of the areal bulk flow to the total area of the ER and the ratio of the areal tubule to the total area of the ER as examples). H: Quantitative and correlation analysis of the ER structural parameters

Fig. 2 Overview of the Blind2Unblind de-noising framework A: Training process. The global masker Ω(·) introduces blind spots into the noisy image y, creating masked patches. Subsequently, the global perc; eptual masking mapper samples h(fθ(Ωy))at the blind spots of the denoised patches. Simultaneously, the denoiser fθ(·) takes y as input and produces the denoised output fθ(y). The re-visible loss employs the imperceptible term h(fθ(Ωy)) as a gradient update medium, facilitating the transition from blind to visible. Additionally, regularization terms are utilized to stabilize the training. B: Inference using the trained denoising model. The denoising network directly generates denoised images from the noisy image y without the need for additional operations.
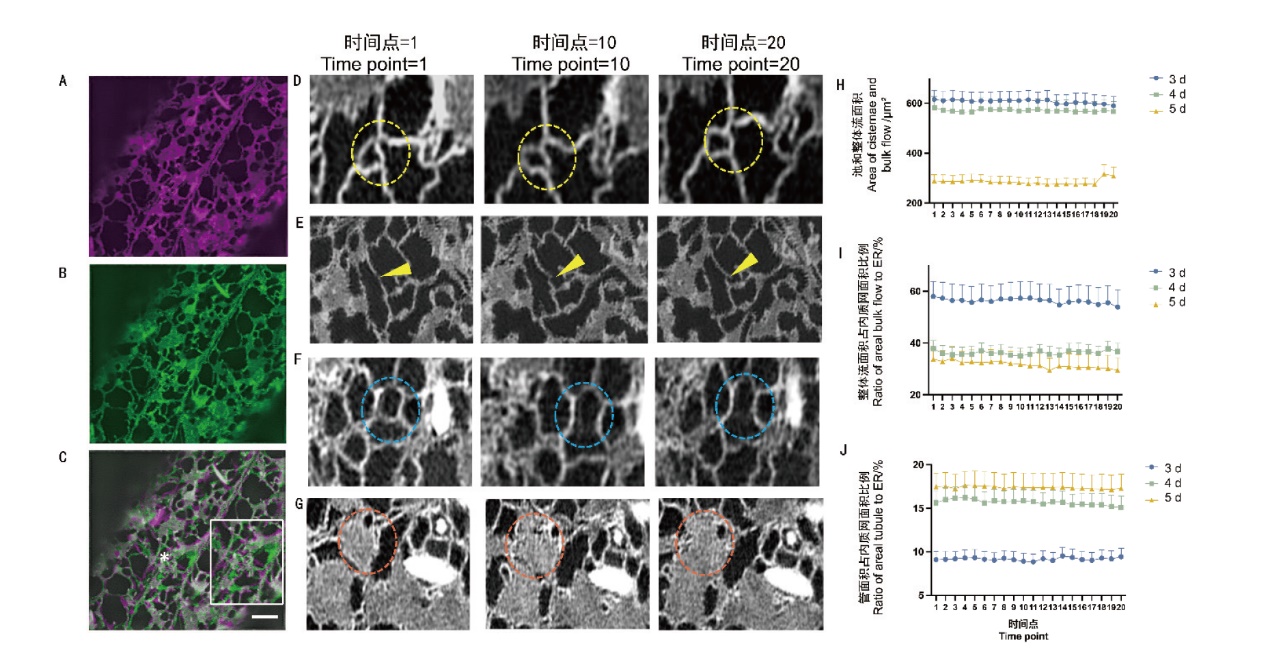
Fig. 3 Structural changes of ER in the time series A: Imaging results of ER structure when the time point of shooting is 0. B: Imaging results of ER structure when the time point of shooting is 20. C: Overlapping results of A and B. The white box on the right side is the demonstrated zoomed the asterisked area. Bar = 2 μm. D: Dynamic change of the three-way junction and multi-way junction inside the circle inside the time series. E: Arrows indicate the dynamics of the growth tip within the time series. F: Dynamic changes of tubules within circles within the time series. G: Dynamic changes of cisternae within circles within the time series. H: Dynamic changes of ER cisternae and bulk flow area under the time series in day 3, 4, and 5. I: Dynamic changes of ER bulk flow area as a proportion of endoplasmic reticulum under the time series of in day 3, 4, and 5. J: Dynamic changes of areal tubule as a proportion of ER area in day 3, 4, and 5 under the time series(n=21 cells)
| 数据 Data | 3 d | 4 d | 5 d | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例 Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | ||||
| 最大值 Maximum | 1 018.63 | 69.76 | 10.98 | 582.88 | 33.67 | 14.27 | 499.43 | 30.70 | 17.13 | |||
| 最小值 Minimum | 236.92 | 66.85 | 9.94 | 32.03 | 32.03 | 13.10 | 486.89 | 27.12 | 16.26 | |||
| 平均数 Mean | 606.85 ±52.46 | 68.71 ±0.78 | 10.38 ±0.06 | 570.93 ±22.58 | 48.71 ±0.72 | 13.65 ±0.12 | 492.25 ±9.92 | 40.36 ±4.26 | 16.79 ±0.05 | |||
Table 1 Statistical data of each ER structural parameter of cells in the elongation zone in the time series
| 数据 Data | 3 d | 4 d | 5 d | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例 Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | ||||
| 最大值 Maximum | 1 018.63 | 69.76 | 10.98 | 582.88 | 33.67 | 14.27 | 499.43 | 30.70 | 17.13 | |||
| 最小值 Minimum | 236.92 | 66.85 | 9.94 | 32.03 | 32.03 | 13.10 | 486.89 | 27.12 | 16.26 | |||
| 平均数 Mean | 606.85 ±52.46 | 68.71 ±0.78 | 10.38 ±0.06 | 570.93 ±22.58 | 48.71 ±0.72 | 13.65 ±0.12 | 492.25 ±9.92 | 40.36 ±4.26 | 16.79 ±0.05 | |||
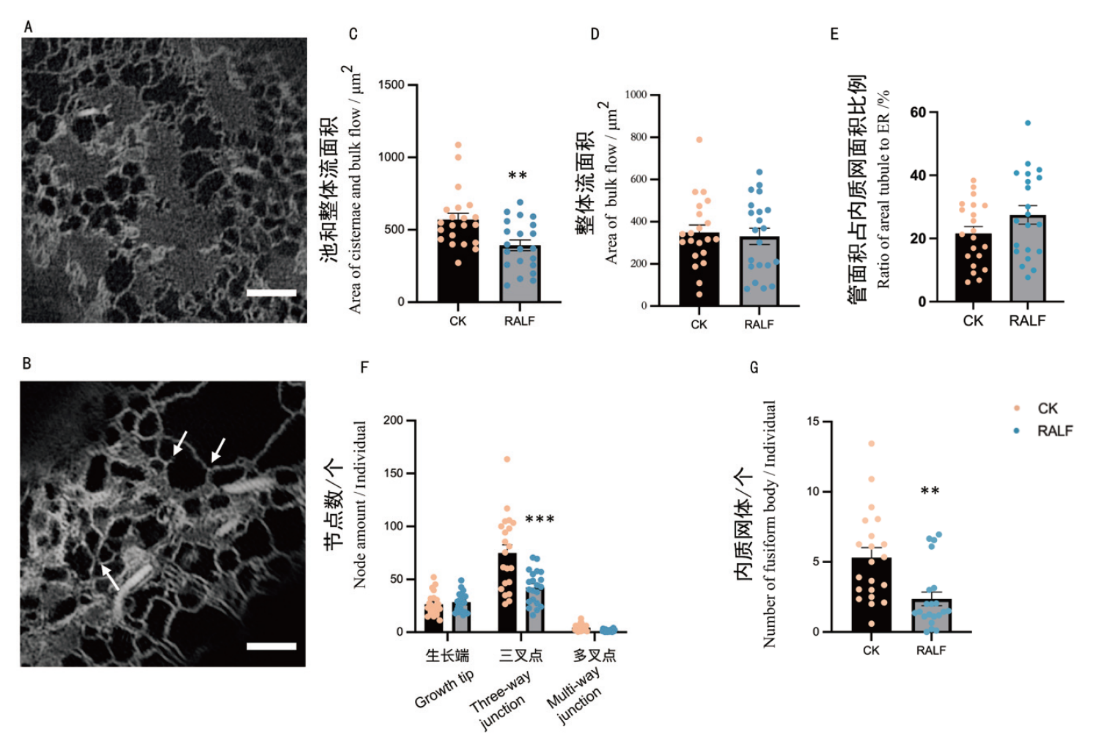
Fig. 4 Elongation zone cells analyzed at day 4 and under RALF stress conditions A: Identification results of elongation zone cells imaged at day 4 under normal growth conditions. B: Identification results of elongation zone cells imaged at day 4 under RALF stress(From left to right, as indicated by the arrows, the sequence is as follows: multi-way junction, growth tip and three-way junction). C: Quantification of areal cisternae and bulk flow under CK and RALF stress. D: Quantification of areal bulk flow under CK and RALF stress. E: Ratio of areal tubule to areal ER under CK and RALF stress quantification. F: Quantification of the number of nodes under CK and RALF stress. G: Quantification of the number of fusiform body under CK and RALF stress. *: P < 0.05, **:P < 0.01, ***: P< 0.001(n=21 cells)
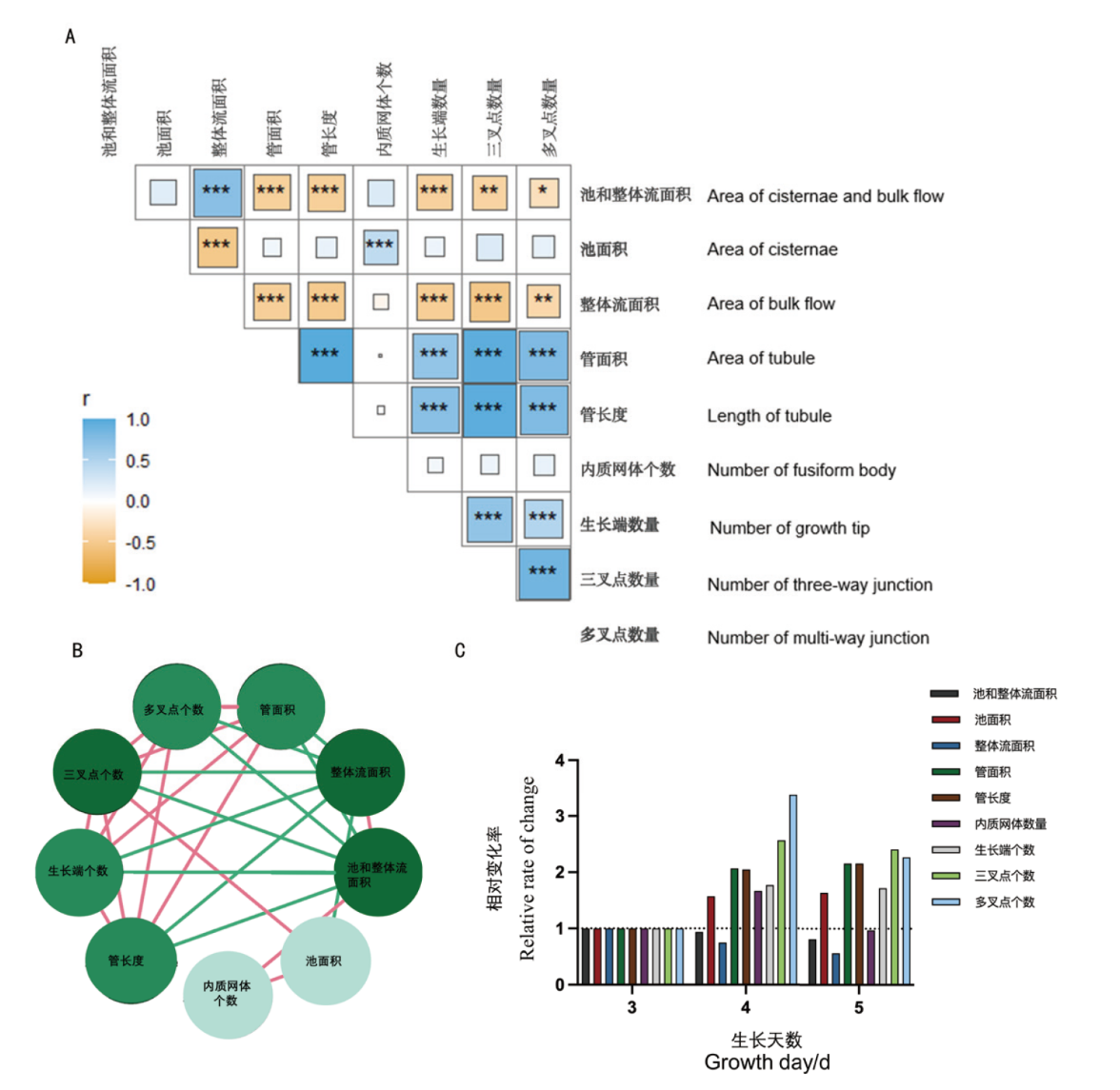
Fig. 5 Comparative correlation analysis of nine feature parameters on the day 3, 4, and 5 of measurement A: Pearson's correlation analysis for quantified data of structural parameters on day 3, 4, and 5, respectively. B: The inter-parameter correlations with red lines denoting positive correlations and green lines representing negative correlations; the intensity of correlation is proportional to the circle color. C: The relative rate of change for the nine feature parameters. *: P< 0.05,**: P< 0.01, ***: P< 0.001
| [1] |
Berridge MJ. The endoplasmic reticulum: a multifunctional signaling organelle[J]. Cell Calcium, 2002, 32(5-6): 235-249.
doi: 10.1016/s0143416002001823 pmid: 12543086 |
| [2] |
Staehelin LA. The plant ER: a dynamic organelle composed of a large number of discrete functional domains[J]. Plant J, 1997, 11(6): 1151-1165.
doi: 10.1046/j.1365-313x.1997.11061151.x pmid: 9225461 |
| [3] | English AR, Voeltz GK. Endoplasmic reticulum structure and interconnections with other organelles[J]. Cold Spring Harb Perspect Biol, 2013, 5(4): a013227. |
| [4] |
Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation[J]. J Cell Biol, 2008, 182(5): 911-924.
doi: 10.1083/jcb.200805140 pmid: 18779370 |
| [5] |
Kadowaki H, Nishitoh H. Endoplasmic reticulum quality control by garbage disposal[J]. FEBS J, 2019, 286(2): 232-240.
doi: 10.1111/febs.14589 pmid: 29923316 |
| [6] |
Agellon LB, Michalak M. The endoplasmic reticulum and the cellular reticular network[J]. Adv Exp Med Biol, 2017, 981: 61-76.
doi: 10.1007/978-3-319-55858-5_4 pmid: 29594858 |
| [7] | Lu M, Christensen CN, Weber JM, et al. ERnet: a tool for the semantic segmentation and quantitative analysis of endoplasmic reticulum topology[J]. Nat Methods, 2023, 20(4): 569-579. |
| [8] |
Bravo R, Parra V, Gatica D, et al. Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration[J]. Int Rev Cell Mol Biol, 2013, 301: 215-290.
doi: 10.1016/B978-0-12-407704-1.00005-1 pmid: 23317820 |
| [9] |
Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains[J]. Int Rev Cytol, 2001, 205: 149-214.
pmid: 11336391 |
| [10] |
Wozny MR, Di Luca A, Morado DR, et al. In situ architecture of the ER-mitochondria encounter structure[J]. Nature, 2023, 618(7963): 188-192.
doi: 10.1038/s41586-023-06050-3 |
| [11] |
Kornmann B, Currie E, Collins SR, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen[J]. Science, 2009, 325(5939): 477-481.
doi: 10.1126/science.1175088 pmid: 19556461 |
| [12] |
Friedman JR, Lackner LL, West M, et al. ER tubules mark sites of mitochondrial division[J]. Science, 2011, 334(6054): 358-362.
doi: 10.1126/science.1207385 pmid: 21885730 |
| [13] |
Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites[J]. Nature, 2013, 495(7441): 389-393.
doi: 10.1038/nature11910 |
| [14] | Li C, Duckney P, Zhang T. et al. TraB family proteins are components of ER-mitochondrial contact sites and regulate ER-mitochondrial interactions and mitophagy[J]. Nat Commun 13,2022, 13(1):5658. |
| [15] |
Ridge RW, Uozumi Y, Plazinski J, et al. Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein[J]. Plant Cell Physiol, 1999, 40(12): 1253-1261.
pmid: 10682347 |
| [16] | Stefano G, Brandizzi F. Unique and conserved features of the plant ER-shaping GTPase RHD3[J]. Cell Logist, 2014, 4(1): e28217. |
| [17] |
Voeltz GK, Prinz WA, Shibata Y, et al. A class of membrane proteins shaping the tubular endoplasmic reticulum[J]. Cell, 2006, 124(3): 573-586.
doi: 10.1016/j.cell.2005.11.047 pmid: 16469703 |
| [18] |
De Craene JO, Coleman J, de Martin PE, et al. Rtn1p is involved in structuring the cortical endoplasmic reticulum[J]. Mol Biol Cell, 2006, 17(7): 3009-3020.
doi: 10.1091/mbc.e06-01-0080 URL |
| [19] |
Tolley N, Sparkes IA, Hunter PR, et al. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport[J]. Traffic, 2008, 9(1): 94-102.
pmid: 17980018 |
| [20] |
West M, Zurek N, Hoenger A, et al. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature[J]. J Cell Biol, 2011, 193(2): 333-346.
doi: 10.1083/jcb.201011039 pmid: 21502358 |
| [21] |
Chang J, Lee S, Blackstone C. Protrudin binds atlastins and endoplasmic reticulum-shaping proteins and regulates network formation[J]. Proc Natl Acad Sci USA, 2013, 110(37): 14954-14959.
doi: 10.1073/pnas.1307391110 pmid: 23969831 |
| [22] | Pain C, Kriechbaumer V, Kittelmann M, et al. Quantitative analysis of plant ER architecture and dynamics[J]. Nat Commun, 2019, 10(1): 984. |
| [23] |
Pearce G, Moura DS, Stratmann J, et al. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development[J]. Proc Natl Acad Sci USA, 2001, 98(22): 12843-12847.
pmid: 11675511 |
| [24] |
Holcman D, Parutto P, Chambers JE, et al. Single particle trajectories reveal active endoplasmic reticulum luminal flow[J]. Nat Cell Biol, 2018, 20(10): 1118-1125.
doi: 10.1038/s41556-018-0192-2 pmid: 30224760 |
| [25] |
Taitt CR, Anderson GP, Ligler FS. Evanescent wave fluorescence biosensors[J]. Biosens Bioelectron, 2005, 20(12):2470-2487.
pmid: 15854820 |
| [26] | Damen D, Hogg DC. Computer vision and pattern recognition(CVPR)[C]. New Orleans:IEEE, 2009 |
| [27] |
Schwarz DS, Blower MD. The endoplasmic reticulum: structure, function and response to cellular signaling[J]. Cell Mol Life Sci, 2016, 73(1): 79-94.
doi: 10.1007/s00018-015-2052-6 pmid: 26433683 |
| [28] | Kim JS, Mochida K, Shinozaki K. ER stress and the unfolded protein response: homeostatic regulation coordinate plant survival and growth[J]. Plants, 2022, 11(23): 3197. |
| [29] | Di Conza G, Ho PC. ER stress responses: an emerging modulator for innate immunity[J]. Cells, 2020, 9(3): 695. |
| [30] |
Zhang LR, Bassham DC, Pittendrigh BR. Editorial: plant ER stress and the UPR signaling pathways[J]. Front Plant Sci, 2022, 13: 968353.
doi: 10.3389/fpls.2022.968353 URL |
| [31] |
Thor F, Gautschi M, Geiger R, et al. Bulk flow revisited: transport of a soluble protein in the secretory pathway[J]. Traffic, 2009, 10(12): 1819-1830.
doi: 10.1111/j.1600-0854.2009.00989.x pmid: 19843282 |
| [1] | SHI Jing-hui, CHEN Wen-hui, LU Kun, ZHENG Ting-ting, REN Zhi-yuan, BAO Guo-qing, WANG Min, LUO Jian-mei. Site-directed Saturation Mutagenesis to Improve the Catalytic Performance of 11α-hydroxylase from Aspergillus ochraceus [J]. Biotechnology Bulletin, 2024, 40(1): 322-331. |
| [2] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [3] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [4] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [5] | LI Yi-ya, WU Yi-fan, DING Neng-shui, FAN Xiao-ping, CHEN Fan. Establishment of a Luciferase-assisted Quantitative Method for Measuring Ultrasonic Disruption of Escherichia coli Cells [J]. Biotechnology Bulletin, 2023, 39(12): 90-98. |
| [6] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [7] | YANG Hua-jie, ZHOU Yu-ping, FAN Tian, LV Tian-xiao, XIE Chu-ping, TIAN Chang-en. Screening and Identification of IQM4-Interacting Proteins in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2021, 37(11): 190-196. |
| [8] | FANG Dan-dan, ZHANG Ting, WEN Xiao-peng. Overexpression of Pinus massoniana PmPT3 Gene in Arabidopsis thaliana Increasing Low Phosphorus Tolerance [J]. Biotechnology Bulletin, 2021, 37(10): 1-8. |
| [9] | XIANG Ming-yuan, LIAO Chang-yu, JIANG Di-ke, ZHANG Peng-fei, WANG Yin, LUO Yan, YANG Ze-xiao, YAO Xue-ping. Establishment and Application of PRRSV NADC30-like SYBR Green I qPCR Detection Method [J]. Biotechnology Bulletin, 2020, 36(5): 80-85. |
| [10] | SUN Xi-lin, JIANG Zhen-yan, LIU Zhi-yi, DAI Lu, SUN Fei, HUANG Wei. Improvement of Thermal Stability of Ganoderma lucidum Protein LZ-8 by Site-directed Mutation of Amino Acids [J]. Biotechnology Bulletin, 2020, 36(1): 23-28. |
| [11] | WANG Qi-wen, LI Pan, Pan Cui-yun, HAN Fen-xia. Effect of Ethylene Glycol on the Expression of Exogenous Genes in Vivo [J]. Biotechnology Bulletin, 2019, 35(4): 64-68. |
| [12] | QI Xin-jie, WANG Yue, WANG Yan-sheng, FANG Guo-kang, HUANG Ying-chun. Applications of Isothermal Titration Calorimetry in Protein-ligand Interactions [J]. Biotechnology Bulletin, 2017, 33(5): 40-49. |
| [13] | XIA Hong-yu KONG Wen-wen XU Rui CANG Wei LI Jing. Improving the Accuracy of FMOGS-OX1 Subcellular Localization Using Cycloheximide [J]. Biotechnology Bulletin, 2017, 33(5): 83-88. |
| [14] | Li Lili, Lin Shuangjun. Recent Advances in ANL Andenylating Enzymes [J]. Biotechnology Bulletin, 2013, 0(4): 14-20. |
| [15] | Wu Wei Liu Chaoqi. Calreticulin and Apoptosis Mediated by Endoplasmic Reticulum Stress [J]. Biotechnology Bulletin, 2013, 29(10): 24-27. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||