








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (1): 322-331.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0751
Previous Articles Next Articles
SHI Jing-hui( ), CHEN Wen-hui, LU Kun, ZHENG Ting-ting, REN Zhi-yuan, BAO Guo-qing, WANG Min, LUO Jian-mei(
), CHEN Wen-hui, LU Kun, ZHENG Ting-ting, REN Zhi-yuan, BAO Guo-qing, WANG Min, LUO Jian-mei( )
)
Received:2023-08-07
Online:2024-01-26
Published:2024-02-06
Contact:
LUO Jian-mei
E-mail:sjh18636348386@163.com;luojianmei@tust.edu.cn
SHI Jing-hui, CHEN Wen-hui, LU Kun, ZHENG Ting-ting, REN Zhi-yuan, BAO Guo-qing, WANG Min, LUO Jian-mei. Site-directed Saturation Mutagenesis to Improve the Catalytic Performance of 11α-hydroxylase from Aspergillus ochraceus[J]. Biotechnology Bulletin, 2024, 40(1): 322-331.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| D118-F | NNKTCTCATGGTTATATTCCTGGTTTTG |
| D118-R | ATCAGTAGTTGGAGTTTCAAAATCCATTC |
| F216-F | NNKGGTGTTGGTGATAAATTGAG |
| F216-R | AGCCAAAGCAGCATATTGAGAAGAAG |
| M488-F | NNKACTTATTTGGCTGATCCAAACAC |
| M488-R | ACCAATATTCAATGGTTGTGGTTTAAAAC |
Table 1 PCR primers for the saturated mutation
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| D118-F | NNKTCTCATGGTTATATTCCTGGTTTTG |
| D118-R | ATCAGTAGTTGGAGTTTCAAAATCCATTC |
| F216-F | NNKGGTGTTGGTGATAAATTGAG |
| F216-R | AGCCAAAGCAGCATATTGAGAAGAAG |
| M488-F | NNKACTTATTTGGCTGATCCAAACAC |
| M488-R | ACCAATATTCAATGGTTGTGGTTTAAAAC |
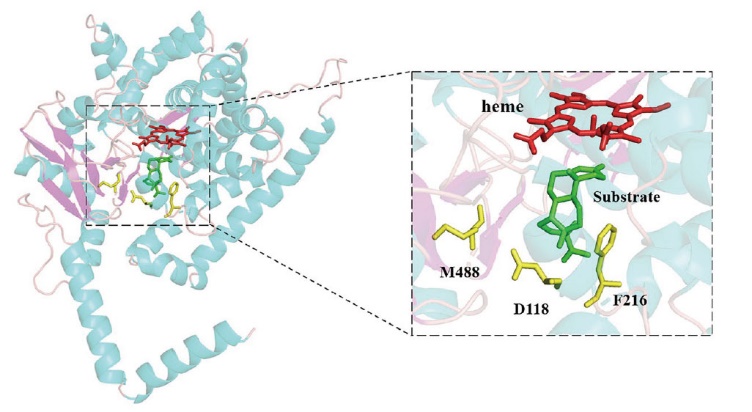
Fig. 1 Distribution of the D118V, F216 and M488 in the docking structure of CYP68J5 with substrate The α-helix, β-folding and random curl of CYP68J5 are colored in bright blue, bright purple and pink, respectively, while the heme, key amino acid sites and substrates are colored in dark red, yellow and green, respectively
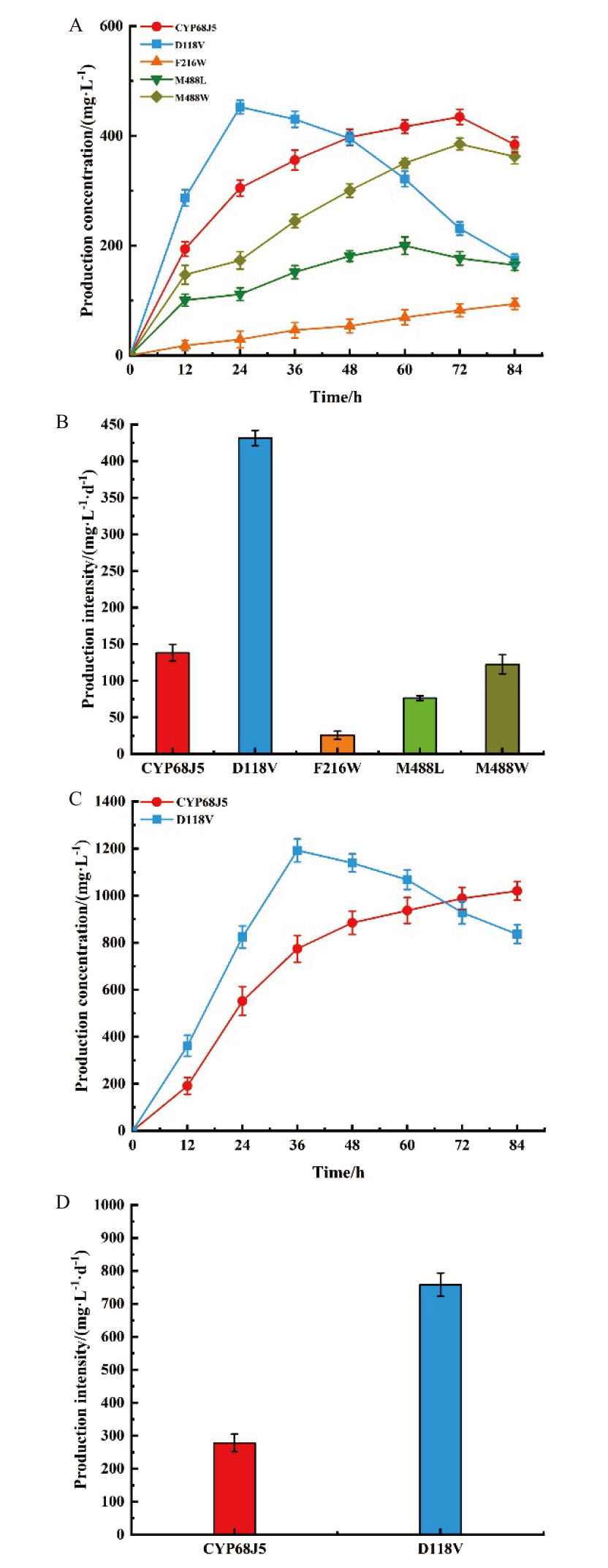
Fig. 3 S. cerevisiae expressing CYP68J5 and its mutants D118V, F216W, M488L, and M488W at 0.5 g/L substrate concentration product formation curves and production intensities(A, B)and 2.0 g/L substrate concentration(C, D)
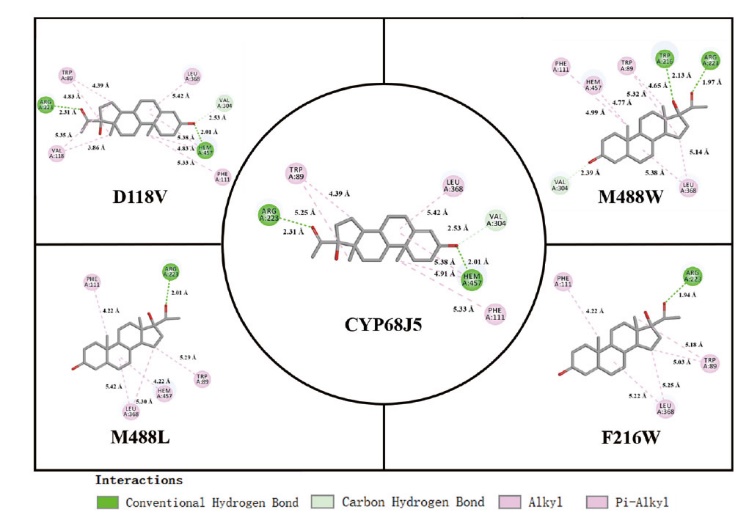
Fig. 4 2D plot of the intermolecular interactions of CYP68J5 and its the mutant D118V, F216W, M488L, and M488W residues and heme with substrates using Discovery Studio

Fig. 5 Molecular dynamics simulations of CYP68J5 and its elite mutant D118V A: Root mean square deviation; B: root mean square fluctuation; C: radius of gyration; D: solvent accessible surface area; E: number of hydrogen bonds; F: free energy
| [1] |
Stouthart AJ, Lucassen EC, van Strien FJ, et al. Stress responsiveness of the pituitary-interrenal axis during early life stages of common carp(Cyprinus carpio)[J]. J Endocrinol, 1998, 157(1): 127-137.
pmid: 9614366 |
| [2] | 张峥斌. 一锅法连续发酵制备Δ1-11α,17α-二羟基黄体酮的方法: CN104711311A[P]. 2015-06-17. |
| Zhang ZB. Preparation of Δ1-11α,17α-dihydroxyprogesterone by continuous fermentation in a one-pot method: 10471131-1A[P]. 2015-06-17. | |
| [3] |
Fotsch C, Wang MH. Blockade of glucocorticoid excess at the tissue level: inhibitors of 11beta-hydroxysteroid dehydrogenase type 1 as a therapy for type 2 diabetes[J]. J Med Chem, 2008, 51(16): 4851-4857.
doi: 10.1021/jm800369f pmid: 18652443 |
| [4] |
Pedersen KB, Geng CD, Vedeckis WV. Three mechanisms are involved in glucocorticoid receptor autoregulation in a human T-lymphoblast cell line[J]. Biochemistry, 2004, 43(34): 10851-10858.
pmid: 15323545 |
| [5] |
Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids—new mechanisms for old drugs[J]. N Engl J Med, 2005, 353(16): 1711-1723.
doi: 10.1056/NEJMra050541 URL |
| [6] | 张喜春, 韩振海, XoджaйoBa ЛТ, 等. 植物体内甾醇的合成和生理作用[J]. 植物生理学通讯, 2001, 37(5): 452-457. |
| Zhang XC, Han ZH, XoджaйoBa ЛT, et al. Synthesis and physiological function of sterols in plants[J]. Plant Physiol Commun, 2001, 37(5): 452-457. | |
| [7] |
Canada KA, Iwashita S, Shim H, et al. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation[J]. J Bacteriol, 2002, 184(2): 344-349.
doi: 10.1128/JB.184.2.344-349.2002 pmid: 11751810 |
| [8] |
Bühler B, Schmid A. Process implementation aspects for biocatalytic hydrocarbon oxyfunctionalization[J]. J Biotechnol, 2004, 113(1-3): 183-210.
pmid: 15380656 |
| [9] |
Urlacher VB, Schmid RD. Recent advances in oxygenase-catalyzed biotransformations[J]. Curr Opin Chem Biol, 2006, 10(2): 156-161.
pmid: 16488653 |
| [10] |
van Beilen JB, Duetz WA, Schmid A, et al. Practical issues in the application of oxygenases[J]. Trends Biotechnol, 2003, 21(4): 170-177.
pmid: 12679065 |
| [11] |
艾露, 陈文慧, 史京辉, 等. 赭曲霉11α羟化酶的克隆表达及关键氨基酸位点分析[J]. 生物技术通报, 2023, 39(4): 114-123.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0572 |
| Ai L, Chen WH, Shi JH, et al. Cloning and expression of 11α hydroxylase from Aspergillus ochraceus and analysis of key amino acid sites[J]. Biotechnol Bull, 2023, 39(4): 114-123. | |
| [12] |
Koehn EM, Kohen A. Flavin-dependent thymidylate synthase: a novel pathway towards thymine[J]. Arch Biochem Biophys, 2010, 493(1): 96-102.
doi: 10.1016/j.abb.2009.07.016 pmid: 19643076 |
| [13] |
Hannemann F, Bichet A, Ewen KM, et al. Cytochrome P450 systems—biological variations of electron transport chains[J]. Biochim Biophys Acta, 2007, 1770(3): 330-344.
pmid: 16978787 |
| [14] |
Barnaba C, Ramamoorthy A. Picturing the membrane-assisted choreography of cytochrome P450 with lipid nanodiscs[J]. Chemphyschem, 2018, 19(20): 2603-2613.
doi: 10.1002/cphc.201800444 pmid: 29995333 |
| [15] | 候向江. 赭曲霉甾体11α-羟化酶基因异源表达研究[D]. 天津: 天津科技大学. |
| Hou XJ. Heterologous expression research of Aspergillus ochraceus steroid 11α-hydrolyase gene[D]. Tianjin: Tianjin University of Science & Technology. | |
| [16] |
Felpeto-Santero C, Galán B, García JL. Production of 11α-hydroxysteroids from sterols in a single fermentation step by Mycolicibacterium smegmatis[J]. Microb Biotechnol, 2021, 14(6): 2514-2524.
doi: 10.1111/1751-7915.13735 pmid: 33660943 |
| [17] | 林本凤, 职亚飞, 刘晓光, 等. 黑曲霉ATCC1015催化16α,17α-环氧黄体酮11α-羟基化及相关P450基因诱导表达[J]. 天津科技大学学报, 2017, 32(6): 8-14. |
| Lin BF, Zhi YF, Liu XG, et al. 11α-hydroxylation of 16α,17α-epoxy progesterone by Aspergillus niger ATCC1015 and induction expression of relevant cytochromes P450 genes[J]. J Tianjin Univ Sci Technol, 2017, 32(6): 8-14. | |
| [18] |
Wang RJ, Sui PC, Hou XJ, et al. Cloning and identification of a novel steroid 11α-hydroxylase gene from Absidia coerulea[J]. J Steroid Biochem Mol Biol, 2017, 171: 254-261.
doi: 10.1016/j.jsbmb.2017.04.006 URL |
| [19] |
Wang X, Yang XW, Jia X, et al. Determination of steroid hydroxylation specificity of an industrial strain Aspergillus ochraceus TCCC41060 by cytochrome P450 gene CYP68J5[J]. Ann Microbiol, 2020, 70(1): 45.
doi: 10.1186/s13213-020-01577-6 |
| [20] |
Qian M, Zeng YL, Mao SH, et al. Engineering of a fungal steroid 11α-hydroxylase and construction of recombinant yeast for improved production of 11α-hydroxyprogesterone[J]. J Biotechnol, 2022, 353: 1-8.
doi: 10.1016/j.jbiotec.2022.05.012 pmid: 35654275 |
| [21] |
Li SL, Chang YW, Liu YN, et al. A novel steroid hydroxylase from Nigrospora sphaerica with various hydroxylation capabilities to different steroid substrates[J]. J Steroid Biochem Mol Biol, 2023, 227: 106236.
doi: 10.1016/j.jsbmb.2022.106236 URL |
| [22] | 乔玉茜. 17α羟基黄体酮11α羟化菌株筛选及其转化工艺研究[D]. 天津: 天津科技大学, 2017. |
| Qiao YQ/X. Screeing and transformation conditions of 11α-hydroxylation of 17α-hydroxy progesterone[D]. Tianjin: Tianjin University of Science & Technology, 2017. | |
| [23] |
Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities[J]. Expert Opin Drug Discov, 2015, 10(5): 449-461.
doi: 10.1517/17460441.2015.1032936 URL |
| [24] | 曲戈, 袁波, 孙周通. 工业蛋白质理性设计与应用[J]. 生物工程学报, 2022, 38(11): 4068-4080. |
| Qu G, Yuan B, Sun ZT. Rational design and applications of industrial proteins[J]. Chin J Biotechnol, 2022, 38(11): 4068-4080. | |
| [25] |
Acevedo-Rocha CG, Gamble CG, Lonsdale R, et al. P450-catalyzed regio- and diastereoselective steroid hydroxylation: efficient directed evolution enabled by mutability landscaping[J]. ACS Catal, 2018, 8(4): 3395-3410.
doi: 10.1021/acscatal.8b00389 URL |
| [26] | Tao S, Gao Y, Li K, et al. Engineering substrate recognition sites of cytochrome P450 monooxygenase CYP116B3 from Rhodococcus ruber for enhanced regiospecific naphthalene hydroxylation[J]. Mol Catal, 2020, 493: 111089. |
| [27] |
Chen J, Fan FY, Qu G, et al. Identification of Absidia orchidis steroid 11β-hydroxylation system and its application in engineering Saccharomyces cerevisiae for one-step biotransformation to produce hydrocortisone[J]. Metab Eng, 2020, 57: 31-42.
doi: 10.1016/j.ymben.2019.10.006 URL |
| [28] |
Doğru EK, Güralp G, Uyar A, et al. Rational design of thermophilic CYP119 for progesterone hydroxylation by in silico mutagenesis and docking screening[J]. J Mol Graph Model, 2023, 118: 108323.
doi: 10.1016/j.jmgm.2022.108323 URL |
| [29] | Tong W, Yan QP, Tian SX, et al. Single-site modification of the P450-BM3 substrate-entrance facilitates the synthesis of optically pure pharmaceutically useful methyl trans-3-phenylglycidates[J]. Mol Catal, 2023, 547: 113354. |
| [30] |
Wang FH, Zhu ML, Song Z, et al. Reshaping the binding pocket of lysine hydroxylase for enhanced activity[J]. ACS Catal, 2020, 10(23): 13946-13956.
doi: 10.1021/acscatal.0c03841 URL |
| [1] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [2] | AI Lu, CHEN Wen-hui, SHI Jing-hui, REN Zhi-yuan, SHEN Wen-qi, YANG Jia-ning, LUO Jian-mei, WANG Min. Cloning and Expression of 11α Hydroxylase from Aspergillus ochraceus and Analysis of Key Amino Acid Sites [J]. Biotechnology Bulletin, 2023, 39(4): 114-123. |
| [3] | HAO Jun-yao, MA Fu-qiang, YANG Guang-yu. Functional Analysis of Key Residues in the Active Center of Creatinase from Alcaligenes sp. KS-85 [J]. Biotechnology Bulletin, 2021, 37(3): 75-83. |
| [4] | QIAO Jing, CUI Sheng-rong, SHI Hong-wu, LUO Zu-liang, MA Xiao-jun. Homology Modeling and Molecular Docking of Cycloartenol Synthase in Siraitia grosvenorii and Speculated Mechanism of Catalytic Cyclization [J]. Biotechnology Bulletin, 2019, 35(2): 101-108. |
| [5] | CHENG Xing-an, YE Jing-min, JIANG Xu-hong, LIU Zhan-mei, Hu Mei-ying. Gene Cloning,Sequence Analysis,and Three Dimensional Structure Prediction of Cathepsin B in Spodoptera frugiperda and Its Molecular Docking Simulation [J]. Biotechnology Bulletin, 2018, 34(1): 183-194. |
| [6] | Wu Congmei, Li Lingling, Guan Xiaoxia, Liu Xintao, Chen Ji, Yin Yuhe. Optimization Screening of Peptides Inhibition to H37Ra [J]. Biotechnology Bulletin, 2014, 0(8): 196-201. |
| [7] | Zhang Guofang, Li Zhenzhen, Yao Weili, Wang Lijun, Zhu Xianming,. High Throughput Drug Screening on Protein PAN of Influenza Virus [J]. Biotechnology Bulletin, 2014, 0(2): 181-186. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||