








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (6): 180-189.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1171
Previous Articles Next Articles
ALIYA Waili1( ), CHEN Yong-kun1, KELAREMU Kelimujiang1, WANG Bao-qing2(
), CHEN Yong-kun1, KELAREMU Kelimujiang1, WANG Bao-qing2( ), CHEN Ling-na1(
), CHEN Ling-na1( )
)
Received:2023-12-13
Online:2024-06-26
Published:2024-05-23
Contact:
WANG Bao-qing, CHEN Ling-na
E-mail:2090926128@qq.com;309114577@qq.com;ln.chen@xjnu.edu.cn
ALIYA Waili, CHEN Yong-kun, KELAREMU Kelimujiang, WANG Bao-qing, CHEN Ling-na. Phylogenetic Evolution and Expression Analysis of SPL Gene Family in Juglans regia[J]. Biotechnology Bulletin, 2024, 40(6): 180-189.
| 软件或在线工具名称Name of the software or online tool | 用途Usage |
|---|---|
| ProtParam在线工具( | 蛋白理化性质预测Prediction of protein physicochemical properties |
| ProtComp9.0( | 亚细胞定位预测Prediction of subcellular localization |
| MEME程序( | 保守基序分析Conservative motif analysis |
| ClustalW2(v2.1) | 多重序列比对Multiple sequence alignment |
| PlantCARE( | 启动子顺式作用元件预测Prediction of promoter cis-acting elements |
Table 1 Bioinformatics software and purpose of usage
| 软件或在线工具名称Name of the software or online tool | 用途Usage |
|---|---|
| ProtParam在线工具( | 蛋白理化性质预测Prediction of protein physicochemical properties |
| ProtComp9.0( | 亚细胞定位预测Prediction of subcellular localization |
| MEME程序( | 保守基序分析Conservative motif analysis |
| ClustalW2(v2.1) | 多重序列比对Multiple sequence alignment |
| PlantCARE( | 启动子顺式作用元件预测Prediction of promoter cis-acting elements |
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | |
|---|---|---|---|---|
| JrSPL1-F | ATCCCCCTCGCAATGGGAGTGG | JrSPL16-R | CCTCCAAAGGCTTGTTCTTCTCCGC | |
| JrSPL1-R | CAGAGAAGCATGTCCCAAGTCTGG | JrSPL20-F | GCAGAGCCGAGGGAAAGAGGAG | |
| JrSPL2-F | ATCCCCCTCGCAATGGGAGTGG | JrSPL20-R | CCGCCTGGCAAGTAATCAGCGTTG | |
| JrSPL2-R | CAGAGAAGCATGTCCCAAGTCTGG | JrSPL21-F | CGCCGATGCCAAGCCGTACC | |
| JrSPL3-F | CTCTCTGGCCGAGTCGGGAG | JrSPL21-R | GTCGCCTCTCATTGTGTCCGGC | |
| JrSPL3-R | CACCACTCCTCACCTTCTTCGGC | JrSPL22-F | CTCTCTTCTGTCATCACCTCCGGC | |
| JrSPL4-F | CGTCTGCTTCTCCACCCATCTCG | JrSPL22-R | CGGATCCCATGTGAAACCTTCCGC | |
| JrSPL4-R | CAGCACACCACCACCCCTCACC | JrSPL23-F | GTGCCAGGTGTGCAGTTGTTGCAAG | |
| JrSPL8-F | ACAGGCCGGATGCCATAGTCGAG | JrSPL23-R | CCCACTTGAATGGGGCAACCGAG | |
| JrSPL8-R | CTGCCTGGCAACATGGCGGTG | JrSPL25-F | GCTATGAGTGCTGCGGATTTGCGG | |
| JrSPL14-F | GGCTCGTGGTGTGGCAGTTCATC | JrSPL25-R | TTGAAGAGCCCCCGGTCAGAGG | |
| JrSPL14-R | CTGAGGGTGTAGGGACTCGGGA | Jractin-F | ATGATGTCAAGGAAGGACTC | |
| JrSPL16-F | GGCACCGCAGCGAATGATGACAC | Jractin-R | CACAATGATCTCAGCTCCG |
Table 2 RT-qPCR primer sequence
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | |
|---|---|---|---|---|
| JrSPL1-F | ATCCCCCTCGCAATGGGAGTGG | JrSPL16-R | CCTCCAAAGGCTTGTTCTTCTCCGC | |
| JrSPL1-R | CAGAGAAGCATGTCCCAAGTCTGG | JrSPL20-F | GCAGAGCCGAGGGAAAGAGGAG | |
| JrSPL2-F | ATCCCCCTCGCAATGGGAGTGG | JrSPL20-R | CCGCCTGGCAAGTAATCAGCGTTG | |
| JrSPL2-R | CAGAGAAGCATGTCCCAAGTCTGG | JrSPL21-F | CGCCGATGCCAAGCCGTACC | |
| JrSPL3-F | CTCTCTGGCCGAGTCGGGAG | JrSPL21-R | GTCGCCTCTCATTGTGTCCGGC | |
| JrSPL3-R | CACCACTCCTCACCTTCTTCGGC | JrSPL22-F | CTCTCTTCTGTCATCACCTCCGGC | |
| JrSPL4-F | CGTCTGCTTCTCCACCCATCTCG | JrSPL22-R | CGGATCCCATGTGAAACCTTCCGC | |
| JrSPL4-R | CAGCACACCACCACCCCTCACC | JrSPL23-F | GTGCCAGGTGTGCAGTTGTTGCAAG | |
| JrSPL8-F | ACAGGCCGGATGCCATAGTCGAG | JrSPL23-R | CCCACTTGAATGGGGCAACCGAG | |
| JrSPL8-R | CTGCCTGGCAACATGGCGGTG | JrSPL25-F | GCTATGAGTGCTGCGGATTTGCGG | |
| JrSPL14-F | GGCTCGTGGTGTGGCAGTTCATC | JrSPL25-R | TTGAAGAGCCCCCGGTCAGAGG | |
| JrSPL14-R | CTGAGGGTGTAGGGACTCGGGA | Jractin-F | ATGATGTCAAGGAAGGACTC | |
| JrSPL16-F | GGCACCGCAGCGAATGATGACAC | Jractin-R | CACAATGATCTCAGCTCCG |

Fig. 4 Phylogenetic relation of SPL family members of J. regia, A. thaliana, S. lycopersicum and P. trichocarpa Sl: S. lycopersicum; Pt: P. trichocarpa; At: A. thaliana; Jr: J. regia
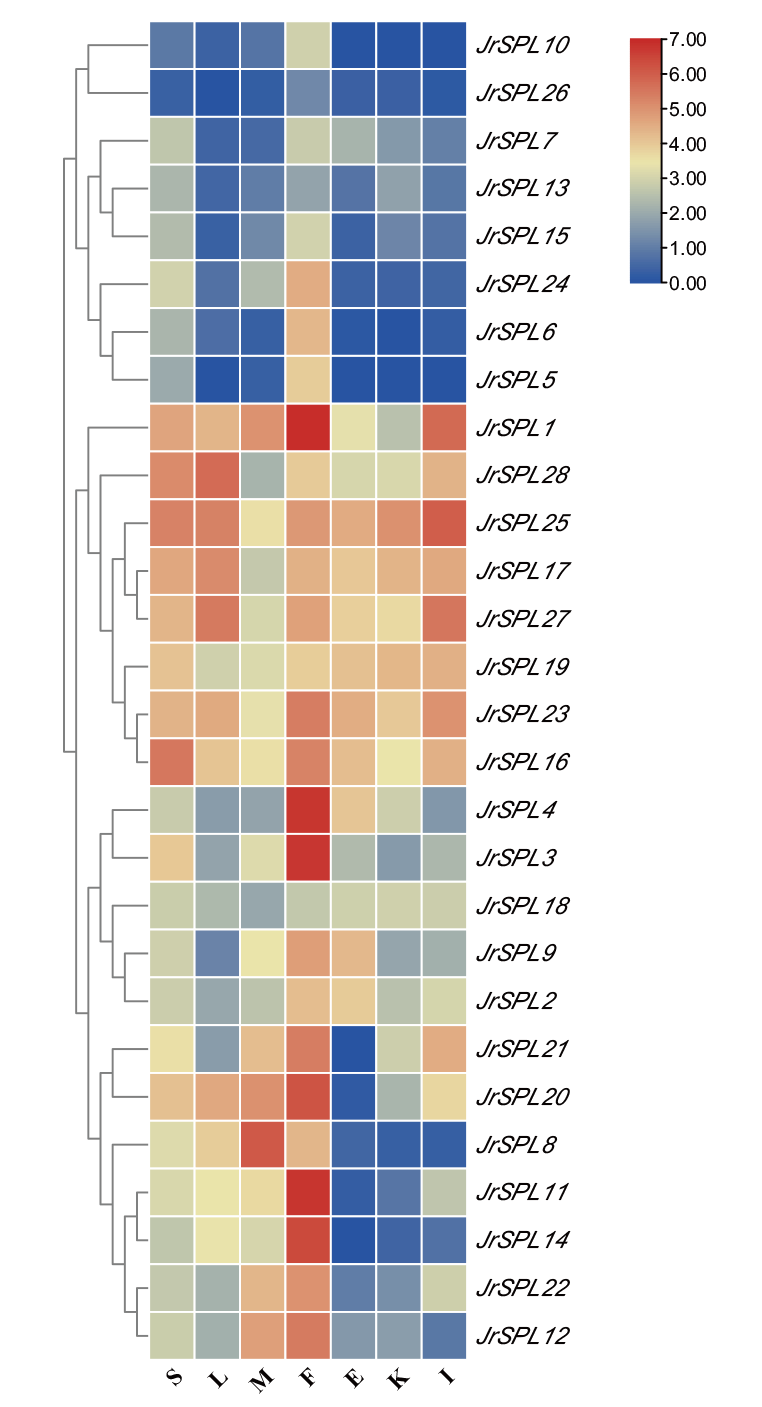
Fig. 5 Expression analysis of JrSPL genes in different tissues The legend indicates the log2 logarithmic transformation on FPKM value. Red indicates high expression levels and blue indicates low expression levels. S: Stem;L: leaf; M: male flower; F: female flower; E: embryo; K: kernel; I: involucre
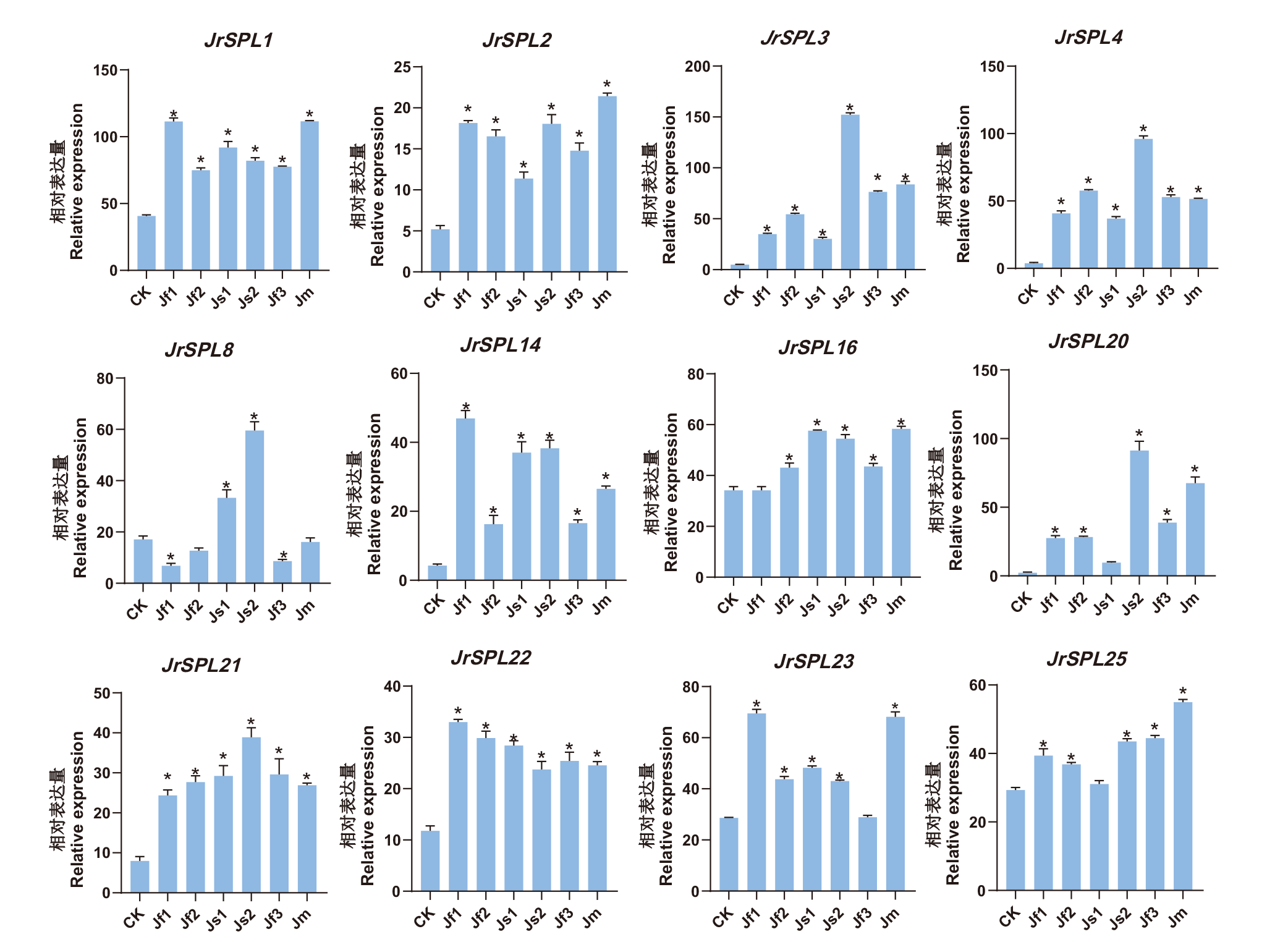
Fig. 7 Relative expression of JrSPL gene in J. regia after grafting-induced flowering CK refers to the stem tips of regenerated branches following the grafting of late fruiting walnut trees. Jf1, Jf2, Jf3, Jm, Js1, and Js2 correspondingly denote distinct floral structures within the flowering material of 'Xinxin 2' post-grafting induction, including pure female flowers (Jf1, Jf2), mixed female flowers (Jf3), male flowers (Jm), and stem tips (Js1, Js2). * indicates a significant difference in relative expression levels compared to the control(P < 0.01)
| [1] | Klein J, Saedler H, Huijser P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA[J]. Mol Gen Genet, 1996, 250(1): 7-16. |
| [2] | 田晶, 赵雪媛, 谢隆聖, 等. SPL转录因子调控植物花发育及其分子机制研究进展[J]. 南京林业大学学报: 自然科学版, 2018, 42(3): 159-166. |
| Tian J, Zhao XY, Xie LS, et al. Research advances and molecular mechanism on SPL transcription factors in regulating plant flower development[J]. J Nanjing For Univ Nat Sci Ed, 2018, 42(3): 159-166. | |
| [3] | Chen XB, Zhang ZL, Liu DM, et al. SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development[J]. J Integr Plant Biol, 2010, 52(11): 946-951. |
| [4] | Yang SM, Overlander-Chen M, Carlson CH, et al. A SQUAMOSA promoter binding protein-like transcription factor controls crop ideotype for high productivity in barley[J]. Plant Direct, 2022, 6(9): e450. |
| [5] | Stief A, Altmann S, Hoffmann K, et al. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors[J]. Plant Cell, 2014, 26(4): 1792-1807. |
| [6] | Guo Q, Li L, Zhao K, et al. Genome-wide analysis of poplar SQUAMOSA-promoter-binding protein(SBP) family under salt stress[J]. Forests, 2021, 12(4): 413. |
| [7] |
Preston JC, Hileman LC. Functional evolution in the plant squamosa-promoter binding protein-like(SPL)gene family[J]. Front Plant Sci, 2013, 4: 80.
doi: 10.3389/fpls.2013.00080 pmid: 23577017 |
| [8] | Li CL, Lu SF. Molecular characterization of the SPL gene family in Populus trichocarpa[J]. BMC Plant Biol, 2014, 14: 131. |
| [9] | Shao FJ, Lu Q, Wilson IW, et al. Genome-wide identification and characterization of the SPL gene family in Ziziphus jujuba[J]. Gene, 2017, 627: 315-321. |
| [10] | He J, Xu ML, Willmann MR, et al. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana[J]. PLoS Genet, 2018, 14(4): e1007337. |
| [11] |
Ma ZB, Li W, Wang HP, et al. WRKY transcription factors WRKY12 and WRKY13 interact with SPL10 to modulate age-mediated flowering[J]. J Integr Plant Biol, 2020, 62(11): 1659-1673.
doi: 10.1111/jipb.12946 |
| [12] | Ma Y, Xue H, Zhang F, et al. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression[J]. Plant Biotechnol J, 2021, 19(2): 311-323. |
| [13] | Yan Y, Wei MX, Li Y, et al. miR529a controls plant height, tiller number, panicle architecture and grain size by regulating SPL target genes in rice(Oryza sativa L.)[J]. Plant Sci, 2021, 302: 110728. |
| [14] | Shikata M, Koyama T, Mitsuda N, et al. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase[J]. Plant Cell Physiol, 2009, 50(12): 2133-2145. |
| [15] |
Manning K, Tör M, Poole M, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening[J]. Nat Genet, 2006, 38(8): 948-952.
doi: 10.1038/ng1841 pmid: 16832354 |
| [16] | Zhou L, Quan SW, Ma L, et al. Molecular characterization of SBP-box gene family during floral induction in walnut(Juglans regia L.)[J]. Tree Genet Genomes, 2019, 16(1): 12. |
| [17] |
Zhang JP, Zhang WT, Ji FY, et al. A high-quality walnut genome assembly reveals extensive gene expression divergences after whole-genome duplication[J]. Plant Biotechnol J, 2020, 18(9): 1848-1850.
doi: 10.1111/pbi.13350 pmid: 32004401 |
| [18] |
Van de Peer Y, Mizrachi E, Marchal K. The evolutionary significance of polyploidy[J]. Nat Rev Genet, 2017, 18: 411-424.
doi: 10.1038/nrg.2017.26 pmid: 28502977 |
| [19] |
冯策婷, 江律, 刘鑫颖, 等. 单叶蔷薇NAC基因家族鉴定及干旱胁迫响应分析[J]. 生物技术通报, 2023, 39(11): 283-296.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0531 |
| Feng CT, Jiang Lyu, Liu XY, et al. Identification of the NAC gene family in Rosa persica and response analysis under drought stress[J]. Biotechnol Bull, 2023, 39(11): 283-296. | |
| [20] |
邢媛, 宋健, 李俊怡, 等. 谷子AP基因家族鉴定及其对非生物胁迫的响应分析[J]. 生物技术通报, 2023, 39(11): 238-251.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0588 |
| Xing Y, Song J, Li JY, et al. Identification of AP gene family and its response analysis to abiotic stress in Setaria italica[J]. Biotechnol Bull, 2023, 39(11): 238-251. | |
| [21] |
Yamaguchi A, Wu MF, Yang L, et al. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1[J]. Dev Cell, 2009, 17(2): 268-278.
doi: 10.1016/j.devcel.2009.06.007 pmid: 19686687 |
| [22] | 李优. 早实核桃花芽分化特点和管理技术要点[J]. 北方园艺, 2015(20): 55. |
| Li Y. Characteristics of flower bud differentiation and management techniques of early fruiting walnut[J]. North Hortic, 2015(20): 55. | |
| [23] | Jung JH, Seo PJ, Kang SK, et al. miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions[J]. Plant Mol Biol, 2011, 76(1/2): 35-45. |
| [1] | GONG Li-li, YU Hua, YANG Jie, CHEN Tian-chi, ZHAO Shuang-ying, WU Yue-yan. Identification and Analysis of Grape(Vitis vinifera L.)CYP707A Gene Family and Functional Verification to Fruit Ripening [J]. Biotechnology Bulletin, 2024, 40(2): 160-171. |
| [2] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [3] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [4] | DUAN Min-jie, LI Yi-fei, YANG Xiao-miao, WANG Chun-ping, HUANG Qi-zhong, HUANG Ren-zhong, ZHANG Shi-cai. Identification of Zinc Finger Protein DnaJ-Like Gene Family in Capsicum annuum and Its Expression Analysis Responses to High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(1): 187-198. |
| [5] | YUAN Xing, GUO Cai-hua, LIU Jin-ming, KANG Chao, QUAN Shao-wen, NIU Jian-xin. Genome-wide Identification of CONSTANS-Like Family Genes and Expression Analysis in Wanlut [J]. Biotechnology Bulletin, 2022, 38(9): 167-179. |
| [6] | CAO Ying-hui, HU Mei-juan, TONG Yan, ZHANG Yan-ping, ZHAO Kai, PENG Dong-hui, ZHOU Yu-zhen. Identification of the ABC Gene Family and Expression Pattern Analysis During Flower Development in Cymbidium ensifolium [J]. Biotechnology Bulletin, 2022, 38(11): 162-174. |
| [7] | GAO Ling, WANG Fei, XIE Shuang-quan, CHEN Xi-feng, SHEN Hai-tao, LI Hong-bin. Genome-wide Identification and Expression Analysis of CBL Gene Family in Glycyrrhiza uralensis [J]. Biotechnology Bulletin, 2021, 37(4): 18-27. |
| [8] | HUANG Ke, HUANG Ge-ge, XUE Man-de, LONG Yan, YUAN Qian-hua, PEI Xin-wu. Cloning and Identification of a Promoter with Root Tissue-specific Expression in Common Wild Rice [J]. Biotechnology Bulletin, 2018, 34(8): 87-92. |
| [9] | ZHAO Zhi-qiang, XUE Man-de, HUANG Ke, ZHANG Jing-wen, LONG Yan, PEI Xin-wu, YUAN Qian-hua. Cloning and Functional Analysis of Green Tissue-specific Expression Promoter of Common Wild Rice(Oryza rufipogon Griff.) [J]. Biotechnology Bulletin, 2017, 33(8): 51-57. |
| [10] | Yan Jun, Guo Xingqi, Cao Xuecheng. Progress of Genome-wide Researches on WRKY Transcription Factors [J]. Biotechnology Bulletin, 2015, 31(11): 9-17. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||