








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 47-52.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0015
Previous Articles Next Articles
LU Xi( ), YUAN Yue, LI Dan, ZHANG Peng(
), YUAN Yue, LI Dan, ZHANG Peng( )
)
Received:2024-01-05
Online:2024-08-26
Published:2024-09-05
Contact:
ZHANG Peng
E-mail:1520624702@qq.com;peng12zhang@gmc.edu.cn
LU Xi, YUAN Yue, LI Dan, ZHANG Peng. Studies on the Regulation of MBD1-induced Expression by Dox in the Tet-On System[J]. Biotechnology Bulletin, 2024, 40(8): 47-52.
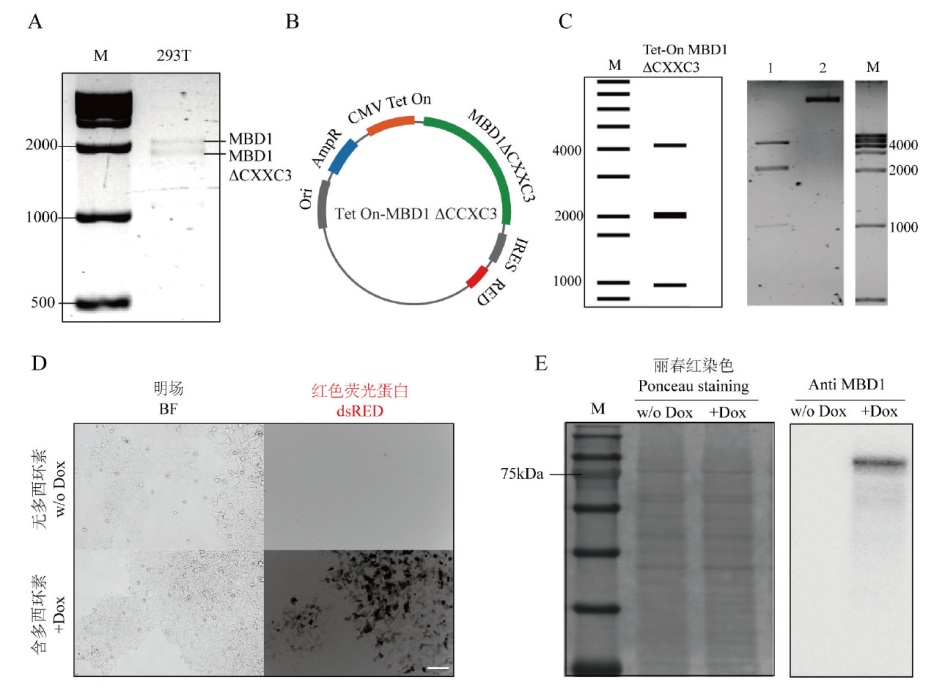
Fig. 1 Construction of Tet-On MBD1ΔCXXC3 plasmid A: The electrophoresis of MBD1 expression sequence amplified from cDNA obtained by RNA reverse transcription in HEK293T; B: the schematic diagram of Tet-On MBD1ΔCXXC3 plasmid construction; C: the electrophoresis diagram of Tet-On MBD1ΔCXXC3 plasmid enzyme digestion, in which M is the 1 000 bp DNA Ladder, and 1 is the restriction nuclease endonuclease BamH I added to the experimental group, and 2 is the negative control group without restriction nucleic acid endonuclease; D: the red fluorescence expression of cells after electrotransformation by Tet-On MBD1ΔCXXC3 plasmid, where the scale bar is 50 μm; E: the MBD1 expression of cells before and after induction by Dox
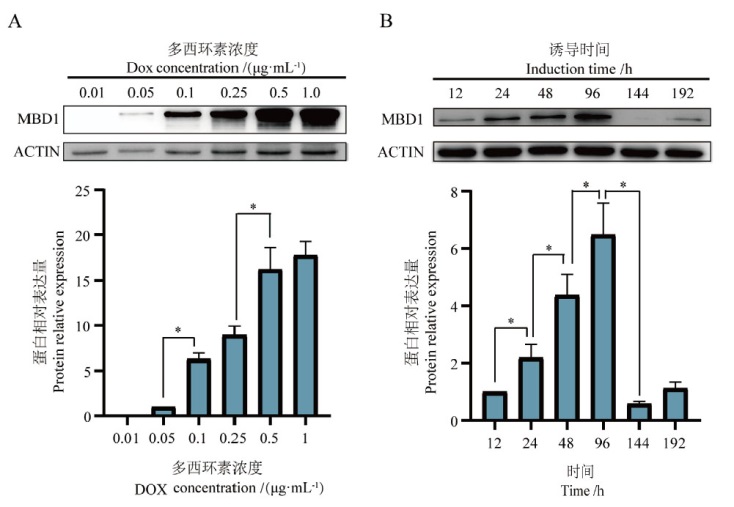
Fig. 2 Effects of Dox on MBD1 expression in Tet-On MBD1 cells A: Expression of MBD1 when induced by different Dox concentrations. B: Expression of MBD1 at different induction times, *P < 0.05. The same below
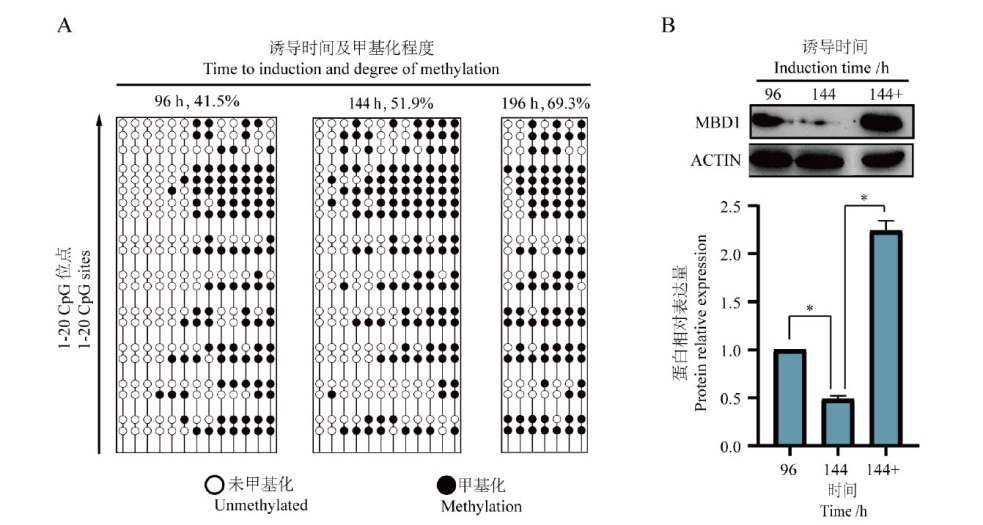
Fig. 3 Effects of Dox induction time on the promoter methylation of Tet-On MBD1ΔCXXC3 plasmid A: Methylation of 20 CpG sites in the promoter region of Tet-On MBD1ΔCXXC3 plasmid under different time of induction; B: expression of MBD1 in the cells after the addition of Decitabine after 96 h of induction by Dox, where 144+ is after 96 h of induction by DOX followed by the addition of Decitabine, and 144- is after 96 h of induction by DOX after DOX induction for 96 h without Decitabine
| [1] |
Navabpour S, Kwapis JL, Jarome TJ. A neuroscientist's guide to transgenic mice and other genetic tools[J]. Neurosci Biobehav Rev, 2020, 108: 732-748.
doi: S0149-7634(19)30610-4 pmid: 31843544 |
| [2] | Kang K, Huang L, Li Q, et al. An improved Tet-on system in microRNA overexpression and CRISPR/Cas9-mediated gene editing[J]. J Anim Sci Biotechnol, 2019, 10: 43. |
| [3] |
Das AT, Tenenbaum L, Berkhout B. Tet-on systems for doxycycline-inducible gene expression[J]. Curr Gene Ther, 2016, 16(3): 156-167.
pmid: 27216914 |
| [4] |
Fan XJ, Petitt M, Gamboa M, et al. Transient, inducible, placenta-specific gene expression in mice[J]. Endocrinology, 2012, 153(11): 5637-5644.
doi: 10.1210/en.2012-1556 pmid: 23011919 |
| [5] | Bertram R, Neumann B, Schuster CF. Status quo of Tet regulation in bacteria[J]. Microb Biotechnol, 2022, 15(4): 1101-1119. |
| [6] | Lin TC, Palei S, Summerer D. Optochemical control of TET dioxygenases enables kinetic insights into the domain-dependent interplay of TET1 and MBD1 while oxidizing and reading 5-methylcytosine[J]. ACS Chem Biol, 2022, 17(7): 1844-1852. |
| [7] |
Gossen M, Freundlieb S, Bender G, et al. Transcriptional activation by tetracyclines in mammalian cells[J]. Science, 1995, 268(5218): 1766-1769.
doi: 10.1126/science.7792603 pmid: 7792603 |
| [8] |
Bertram R, Hillen W. The application of Tet repressor in prokaryotic gene regulation and expression[J]. Microb Biotechnol, 2008, 1(1): 2-16.
doi: 10.1111/j.1751-7915.2007.00001.x pmid: 21261817 |
| [9] |
Loew R, Heinz N, Hampf M, et al. Improved Tet-responsive promoters with minimized background expression[J]. BMC Biotechnol, 2010, 10: 81.
doi: 10.1186/1472-6750-10-81 pmid: 21106052 |
| [10] | O'Donnell KA, An WF, Schrum CT, et al. Controlled insertional mutagenesis using a LINE-1(ORFeus)gene-trap mouse model[J]. Proc Natl Acad Sci USA, 2013, 110(29): E2706-E2713. |
| [11] |
Ali Hosseini Rad SM, Poudel A, Tan GMY, et al. Optimisation of Tet-On inducible systems for Sleeping Beauty-based chimeric antigen receptor(CAR)applications[J]. Sci Rep, 2020, 10(1): 13125.
doi: 10.1038/s41598-020-70022-0 pmid: 32753634 |
| [12] | Wanka F, Cairns T, Boecker S, et al. Tet-on, or Tet-off, that is the question: advanced conditional gene expression in Aspergillus[J]. Fungal Genet Biol, 2016, 89: 72-83. |
| [13] |
Allan AM, Liang XM, Luo YP, et al. The loss of methyl-CpG binding protein 1 leads to autism-like behavioral deficits[J]. Hum Mol Genet, 2008, 17(13): 2047-2057.
doi: 10.1093/hmg/ddn102 pmid: 18385101 |
| [14] | Saunderson SC, Hosseini-Rad SMA, McLellan AD. Noise-reduction and sensitivity-enhancement of a sleeping beauty-based tet-on system[J]. Genes, 2022, 13(10): 1679. |
| [15] | Benati D, Cocchiarella F, Recchia A. An efficient in vitro transposition method by a transcriptionally regulated sleeping beauty system packaged into an integration defective lentiviral vector[J]. J Vis Exp, 2018(131): 56742. |
| [16] | Hubner EK, Lechler C, Kohnke-Ertel B, et al. An in vivo transfection system for inducible gene expression and gene silencing in murine hepatocytes[J]. J Gene Med, 2017, 19(1-2): 10.1002/jgm.2940. |
| [17] |
Geurts AM, Wilber A, Carlson CM, et al. Conditional gene expression in the mouse using a Sleeping Beauty gene-trap transposon[J]. BMC Biotechnol, 2006, 6: 30.
pmid: 16800892 |
| [18] |
Meng H, Cao Y, Qin JZ, et al. DNA methylation, its mediators and genome integrity[J]. Int J Biol Sci, 2015, 11(5): 604-617.
doi: 10.7150/ijbs.11218 pmid: 25892967 |
| [19] |
Clouaire T, de Las Heras JI, Merusi C, et al. Recruitment of MBD1 to target genes requires sequence-specific interaction of the MBD domain with methylated DNA[J]. Nucleic Acids Res, 2010, 38(14): 4620-4634.
doi: 10.1093/nar/gkq228 pmid: 20378711 |
| [20] |
Fujita N, Takebayashi S, Okumura K, et al. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms[J]. Mol Cell Biol, 1999, 19(9): 6415-6426.
doi: 10.1128/MCB.19.9.6415 pmid: 10454587 |
| [21] | Li L, Chen BF, Chan WY. An epigenetic regulator: methyl-CpG-binding domain protein 1(MBD1)[J]. Int J Mol Sci, 2015, 16(3): 5125-5140. |
| [1] | YUAN Ming-bo, YE Guo-hua, YANG Dan, SONG Dong-xue. Research Progress in DNA Methylation Sequencing Technology [J]. Biotechnology Bulletin, 2024, 40(5): 58-65. |
| [2] | LI Ying, YUE Xiang-hua. Application of DNA Methylation in Interpreting Natural Variation in Moso Bamboo [J]. Biotechnology Bulletin, 2023, 39(7): 48-55. |
| [3] | AN Lei, ZHAO Jin-ling, REN Xiao-liang. RNA Modification and Its Research Progress in Caenorhabditis elegans [J]. Biotechnology Bulletin, 2023, 39(4): 176-186. |
| [4] | ZHANG Miao, YANG Lu-lu, JIA Yan-long, WANG Tian-yun. Research Progress in the Roles of DNA and Histone Methylations in Epigenetic Regulation [J]. Biotechnology Bulletin, 2022, 38(7): 23-30. |
| [5] | WANG Chen-chen, ZHANG Fan-li, CHEN Pei-qi, WENG Si-yao, WANG Hui-fang, CUI Xiao-juan. Research Progress in the Structural and Functional Analysis of Mammalian DNA Methyltransferase DNMT1 and DNMT3 [J]. Biotechnology Bulletin, 2022, 38(7): 31-39. |
| [6] | WANG Jian-yong, ZOU Yong-mei, GE Yan-bin, WANG Kai, XI Meng-li. Advance on Epigenetic Modification During Plant Callus Induction [J]. Biotechnology Bulletin, 2021, 37(8): 253-262. |
| [7] | TANG De-ping, YAO Hui-hui, TANG Jin-zhou, MAO Ai-hong. Mutual Regulation of microRNAs and Epigenetics in Human Cancers [J]. Biotechnology Bulletin, 2020, 36(8): 194-200. |
| [8] | WANG Jie, XIE Li-nan. Research Progress of Demethylase ROS1 in Plants [J]. Biotechnology Bulletin, 2020, 36(7): 148-157. |
| [9] | LIU Zhi-min, YANG Zhi-yi, JI Feng-dan, MEI Zhi-chao, YU Jia-hui, XIE Li-nan. Research Progress of Plant DNA Methylation Under Abiotic Stress [J]. Biotechnology Bulletin, 2020, 36(11): 122-132. |
| [10] | JIANG Rui, LÜ Ke-nao, PAN Xue-feng, CUI Xin-xia, SHEN Shi-gang, DING Liang. Current Status and Challenges of Epigenetic Drug Research and Development [J]. Biotechnology Bulletin, 2019, 35(8): 213-225. |
| [11] | LONG Wen-lin, GUO Hui, SHENG Jie, SONG Ru-hui, XU Yao. Role of m6A RNA Methylation in Tumorigenesis and Development [J]. Biotechnology Bulletin, 2019, 35(6): 178-186. |
| [12] | XUE Jing-jing, CHEN Song-bi. Variation Analysis of DNA Methylation in Different Development Stages of Cassava [J]. Biotechnology Bulletin, 2018, 34(5): 117-123. |
| [13] | AO Xu-dong ,SA Ru-la, WANG Jie, WANG Hui-min ,YU Hai-quan. The Effect of DNA Methyltransferase Inhibitor 5-Aza-CdR on AID Gene-modified Bovine Fetal Fibroblasts [J]. Biotechnology Bulletin, 2016, 32(8): 103-112. |
| [14] | AO Xu-dong SA Ru-la WANG Jie WANG Hui-min YU Hai-quan. Expression of AID and Dynamic Changes of Its DNA Methylation in Regulation Region During Bovine Early Embryonic Development [J]. Biotechnology Bulletin, 2016, 32(7): 242-249. |
| [15] | ZHAO Qian, WANG Wei ,SUN Ye-qing. Review on the Correlation between DNA Methylation and Gene Expression in Plant [J]. Biotechnology Bulletin, 2016, 32(4): 16-23. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||