








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (11): 227-235.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0318
Previous Articles Next Articles
WANG Qi-xin1( ), ZHANG Ling-xin1, ZHONG Ni-na1, XIONG Wang-dan1,2(
), ZHANG Ling-xin1, ZHONG Ni-na1, XIONG Wang-dan1,2( )
)
Received:2024-03-31
Online:2024-11-26
Published:2024-12-19
Contact:
XIONG Wang-dan
E-mail:2730926732@qq.com;xiongwd@qau.edu.cn
WANG Qi-xin, ZHANG Ling-xin, ZHONG Ni-na, XIONG Wang-dan. Identification and Expression Analysis of ARF Gene Family in Setaria viridis[J]. Biotechnology Bulletin, 2024, 40(11): 227-235.
| 基因名称Gene name | 正向引物 Forword primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| SvARF5 | GCAGTTCCGTCACATCTACCG | CGAGGAGTCATCCCAGTCAA |
| SvARF6 | ACCCGTGGGAGTCATTTGTG | GATAGGTAGCGTGGATCGTTTC |
| SvARF9 | AATTGGCTCGTTCGATTACG | ACTTGCCCTTGGCTGCTGT |
| SvARF10 | TATTTGGGTTTCCTCTTGGTAG | TTCTGAACATTCTGGGTAGCCT |
| SvARF14 | CCCTTCTTGCTCAACATCACC | CTTCGGCAGTTCCTTAGTCG |
| SvARF17 | TTCTCCAAAGCCTGAGCAAT | CAAATCTTGGAACCCCGAAG |
| SvARF18 | TGATGGTGAACTAAGACTGGGTG | AGACTGGACGGTTGAGGCTC |
| SvARF22 | CCTGACCTCTGGCACCCTGAT | TCCCAAGCACAAGCACCCAC |
| SvARF24 | GGTTTCTCATCTCAAGGTCGTG | AGCCTCCGTGCTACTTTCAT |
| SvActin | CAGTGGACGCACAACAGGTAT | AGCAAGGTCAAGACGGAGAAT |
Table 1 Primer sequences for quantitative real time PCR
| 基因名称Gene name | 正向引物 Forword primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| SvARF5 | GCAGTTCCGTCACATCTACCG | CGAGGAGTCATCCCAGTCAA |
| SvARF6 | ACCCGTGGGAGTCATTTGTG | GATAGGTAGCGTGGATCGTTTC |
| SvARF9 | AATTGGCTCGTTCGATTACG | ACTTGCCCTTGGCTGCTGT |
| SvARF10 | TATTTGGGTTTCCTCTTGGTAG | TTCTGAACATTCTGGGTAGCCT |
| SvARF14 | CCCTTCTTGCTCAACATCACC | CTTCGGCAGTTCCTTAGTCG |
| SvARF17 | TTCTCCAAAGCCTGAGCAAT | CAAATCTTGGAACCCCGAAG |
| SvARF18 | TGATGGTGAACTAAGACTGGGTG | AGACTGGACGGTTGAGGCTC |
| SvARF22 | CCTGACCTCTGGCACCCTGAT | TCCCAAGCACAAGCACCCAC |
| SvARF24 | GGTTTCTCATCTCAAGGTCGTG | AGCCTCCGTGCTACTTTCAT |
| SvActin | CAGTGGACGCACAACAGGTAT | AGCAAGGTCAAGACGGAGAAT |
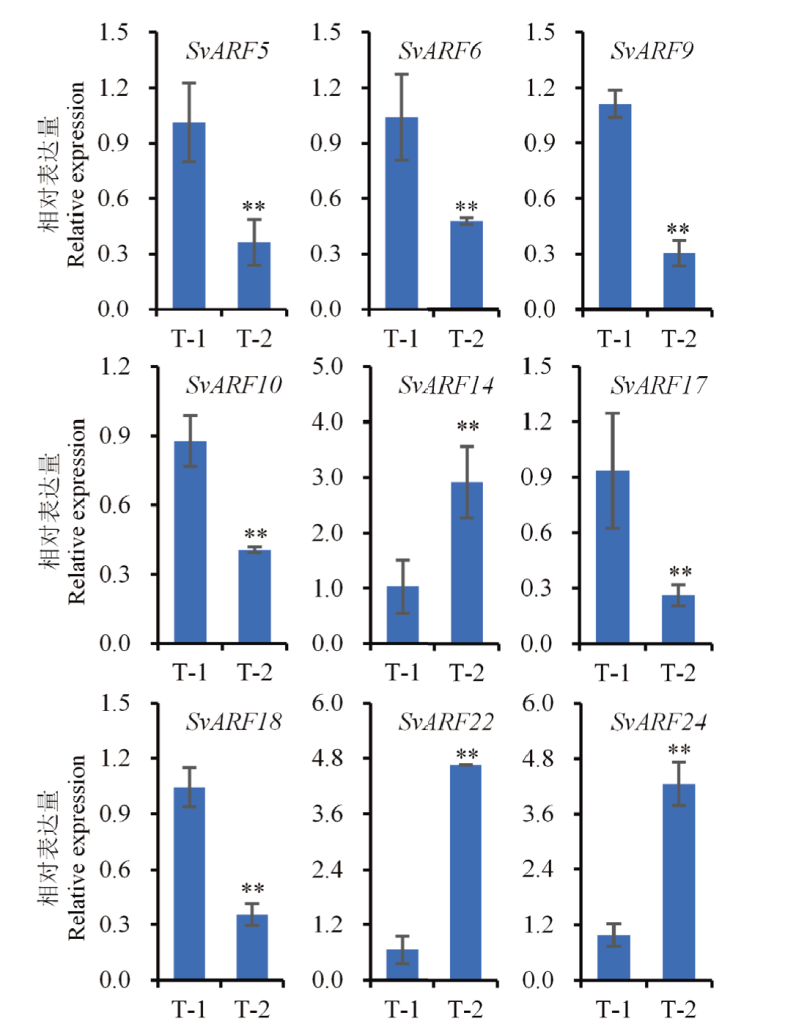
Fig. 7 Relative expression analysis of SvARF genes in the tillers of S. viridis after spraying 2,4-D isooctyl ester The expression of SvARF genes under control T0-1 was defined as 1, and the relative expression of SvARF gene under treatment was determined by the relative fold changes between treatment and control. T-1 and T-2 refer to the concentration of 2,4-D isooctyl ester at 1 050 and 4 200 g a.i./ha, respectively. ** indicates P<0.01. The same below

Fig. 8 Content of IAA in the tillers of S. viridis after spraying 2,4-D isooctyl ester T-0 refers to the concentration of 2,4-D isooctyl ester at 0 g a.i./ha
| [1] | Wang JH, Hu D, Shi XN, et al. Different responses of invasive weed Alternanthera philoxeroides and Oryza sativa to plant growth regulators[J]. Life, 2022, 12(7): 1069. |
| [2] |
Guo F, Huang YZ, Qi PP, et al. Functional analysis of auxin receptor OsTIR1/OsAFB family members in rice grain yield, tillering, plant height, root system, germination, and auxinic herbicide resistance[J]. New Phytol, 2021, 229(5): 2676-2692.
doi: 10.1111/nph.17061 pmid: 33135782 |
| [3] |
Ishikawa S, Maekawa M, Arite T, et al. Suppression of tiller bud activity in tillering dwarf mutants of rice[J]. Plant Cell Physiol, 2005, 46(1): 79-86.
doi: 10.1093/pcp/pci022 pmid: 15659436 |
| [4] | Yu HY, Cui HL, Chen JC, et al. Regulation of 2, 4-D isooctyl ester on Triticum aestivum and Aegilops tauschii tillering and endogenous phytohormonal responses[J]. Front Plant Sci, 2021, 12: 642701. |
| [5] |
Roosjen M, Paque S, Weijers D. Auxin Response Factors: output control in auxin biology[J]. J Exp Bot, 2018, 69(2): 179-188.
doi: 10.1093/jxb/erx237 pmid: 28992135 |
| [6] |
Dinesh DC, Kovermann M, Gopalswamy M, et al. Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response[J]. Proc Natl Acad Sci USA, 2015, 112(19): 6230-6235.
doi: 10.1073/pnas.1424077112 pmid: 25918389 |
| [7] |
Li YH, Han S, Qi YH. Advances in structure and function of auxin response factor in plants[J]. J Integr Plant Biol, 2023, 65(3): 617-632.
doi: 10.1111/jipb.13392 |
| [8] |
Wilmoth JC, Wang SC, Tiwari SB, et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation[J]. Plant J, 2005, 43(1): 118-130.
doi: 10.1111/j.1365-313X.2005.02432.x pmid: 15960621 |
| [9] |
Yang J, Tian L, Sun MX, et al. AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis[J]. Plant Physiol, 2013, 162(2): 720-731.
doi: 10.1104/pp.113.214940 pmid: 23580594 |
| [10] | Ellis CM, Nagpal P, Young JC, et al. AUXIN RESPONSE FACTOR 1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana[J]. Development, 2005, 132(20): 4563-4574. |
| [11] | Sims K, Abedi-Samakush F, Szulc N, et al. OsARF11 promotes growth, meristem, seed, and vein formation during rice plant development[J]. Int J Mol Sci, 2021, 22(8): 4089. |
| [12] | Li PH, Brutnell TP. Setaria viridis and Setaria italica, model genetic systems for the Panicoid grasses[J]. J Exp Bot, 2011, 62(9): 3031-3037. |
| [13] |
Keshavarzi M, Seifali M. Systematic study of weedy species of Setaria(L.) P. Beauv.(Poaceae)in Iran[J]. Pak J Biol Sci, 2007, 10(19): 3362-3367.
pmid: 19090151 |
| [14] | Xiong WD, Xu XQ, Zhang L, et al. Genome-wide analysis of the WRKY gene family in physic nut(Jatropha curcas L.)[J]. Gene, 2013, 524(2): 124-132. |
| [15] | Zhao S, Zhang ML, Ma TL, et al. Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress[J]. Plant Cell, 2016, 28(12): 3005-3019. |
| [16] | Chen Y, Liu B, Zhao YJ, et al. Whole-genome duplication and purifying selection contributes to the functional redundancy of auxin response factor(ARF)genes in foxtail millet(Setaria italica L.)[J]. Int J Genomics, 2021, 2021: 2590665. |
| [17] | Wang DK, Pei KM, Fu YP, et al. Genome-wide analysis of the auxin response factors(ARF)gene family in rice(Oryza sativa)[J]. Gene, 2007, 394(1/2): 13-24. |
| [18] | Song H, Guo ZL, Zhang XJ, et al. De novo genes in Arachis hypogaea cv. Tifrunner: systematic identification, molecular evolution, and potential contributions to cultivated peanut[J]. Plant J, 2022, 111(4): 1081-1095. |
| [19] | Liu R, Guo ZF, Lu SY. Genome-wide identification and expression analysis of the Aux/IAA and auxin response factor gene family in Medicago truncatula[J]. Int J Mol Sci, 2021, 22(19): 10494. |
| [20] | Chen FQ, Zhang JQ, Ha X, et al. Genome-wide identification and expression analysis of the auxin-response factor(ARF)gene family in Medicago sativa under abiotic stress[J]. BMC Genomics, 2023, 24(1): 498. |
| [21] |
Jiao YN. Double the genome, double the fun: genome duplications in angiosperms[J]. Mol Plant, 2018, 11(3): 357-358.
doi: S1674-2052(18)30059-5 pmid: 29476919 |
| [22] | Peng YK, Zhao K, Zheng RY, et al. A comprehensive analysis of auxin response factor gene family in Melastoma dodecandrum genome[J]. Int J Mol Sci, 2024, 25(2): 806. |
| [23] |
Okushima Y, Overvoorde PJ, Arima K, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19[J]. Plant Cell, 2005, 17(2): 444-463.
doi: 10.1105/tpc.104.028316 pmid: 15659631 |
| [24] | Wang HX, Ren J, Zhou SY, et al. Molecular regulation of oil gland development and biosynthesis of essential oils in Citrus spp[J]. Science, 2024, 383(6683): 659-666. |
| [25] |
Mockaitis K, Estelle M. Auxin receptors and plant development: a new signaling paradigm[J]. Annu Rev Cell Dev Biol, 2008, 24: 55-80.
doi: 10.1146/annurev.cellbio.23.090506.123214 pmid: 18631113 |
| [26] |
Korasick DA, Jez JM, Strader LC. Refining the nuclear auxin response pathway through structural biology[J]. Curr Opin Plant Biol, 2015, 27: 22-28.
doi: 10.1016/j.pbi.2015.05.007 pmid: 26048079 |
| [27] | Chandler JW. Auxin response factors[J]. Plant Cell Environ, 2016, 39(5): 1014-1028. |
| [28] |
Grossmann K. Auxin herbicides: current status of mechanism and mode of action[J]. Pest Manag Sci, 2010, 66(2): 113-120.
doi: 10.1002/ps.1860 pmid: 19823992 |
| [29] | Yan M, Yan Y, Wang P, et al. Genome-wide identification and expression analysis of auxin response factor(ARF)gene family in Panax ginseng indicates its possible roles in root development[J]. Plants, 2023, 12(23): 3943. |
| [30] | Li Y, Li JL, Chen ZH, et al. OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice[J]. Plant Biotechnol J, 2020, 18(10): 2015-2026. |
| [31] | Li JC, Jiang YM, Zhang J, et al. Key auxin response factor(ARF)genes constraining wheat tillering of mutant dmc[J]. Peer J, 2021, 9: e12221. |
| [32] |
Su LY, Zhang T, Yang B, et al. Different evolutionary patterns of TIR1/AFBs and AUX/IAAs and their implications for the morphogenesis of land plants[J]. BMC Plant Biol, 2023, 23(1): 265.
doi: 10.1186/s12870-023-04253-4 pmid: 37202746 |
| [1] | ZHANG Man-yu, DONG Jia-cheng, GOU Fu-fan, GONG Chao-hui, LIU Qian, SUN Wen-liang, KONG zhen, HAO Jie, WANG Min, TIAN Chao-guang. Cloning, Expression, Characterization and Application of the Pectin Esterase MtCE12-1 from Myceliophthora thermophila [J]. Biotechnology Bulletin, 2024, 40(9): 291-300. |
| [2] | WANG Qian, ZHOU Jia-yan, WANG Qian, DENG Yu-ping, ZHANG Min-hui, CHEN Jing, YANG Jun, ZOU Jian. Identification and Expression Analysis of the YABBY Gene Family in Sunflower [J]. Biotechnology Bulletin, 2024, 40(8): 199-211. |
| [3] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [4] | REN Xiao-min, YUN Lan, AI Qian, ZHAO Qiao. Functional Verification of Isopentenyl Transferases PjIPT Gene in Psathyrostachys juncea [J]. Biotechnology Bulletin, 2024, 40(7): 207-215. |
| [5] | QIN Jian, LI Zhen-yue, HE Lang, LI Jun-ling, ZHANG Hao, DU Rong. Change of Single-cell Transcription Profile and Analysis of Intercellular Communication in Myogenic Cell Differentiation [J]. Biotechnology Bulletin, 2024, 40(6): 330-342. |
| [6] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| [7] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| [8] | CHEN Qiang, HUANG Xin-hui, ZHANG Zheng, ZHANG Chong, LIU Ye-fei. Effects of Melatonin on the Fruit Softening and Ethylene Synthesis of Post-harvest Oriental Melon [J]. Biotechnology Bulletin, 2024, 40(4): 139-147. |
| [9] | ZHANG Yu, SHI Lei, GONG Lei, NIE Feng-jie, YANG Jiang-wei, LIU Xuan, YANG Wen-jing, ZHANG Guo-hui, XIE Rui-xia, ZHANG Li. Genome-wide Identification of Potato WOX Gene Family and Its Expression Analysis in in vitro Regeneration and Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(3): 170-180. |
| [10] | WU Xing-xing, HONG Hai-bo, GAN Zhi-cheng, LI Rui-ning, HUANG Xian-zhong. Cloning and Preliminary Functional Analysis of CaPI Gene in Capsicum annuum L. [J]. Biotechnology Bulletin, 2024, 40(3): 193-201. |
| [11] | JIANG Lin-qi, ZHAO Jia-ying, ZHENG Fei-xiong, YAO Xin-yi, LI Xiao-xian, YU Zhen-ming. Identification and Expression Analysis of 14-3-3 Gene Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2024, 40(3): 229-241. |
| [12] | WANG Feng-ting, ZHAO Fu-shun, QIAO Kai-bin, XU Xun, LIU Jin-liang. Progress on the Molecular Mechanism of Scion-rootstock Interactions in Vegetable Grafting [J]. Biotechnology Bulletin, 2024, 40(10): 149-159. |
| [13] | WU Zhen, ZHANG Ming-Ying, YAN Feng, LI Yi-min, GAO Jing, YAN Yong-Gang, ZHANG Gang. Identification and Analysis of WRKY Gene Family in Rheum palmatum L. [J]. Biotechnology Bulletin, 2024, 40(1): 250-261. |
| [14] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [15] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||