








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (1): 198-209.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0596
Previous Articles Next Articles
LI Yun-bin1( ), LI Yu-ping1, CHENG Qin1, LIN Chun1,2, MAO Zi-chao1,2, LIU Zheng-jie1,2(
), LI Yu-ping1, CHENG Qin1, LIN Chun1,2, MAO Zi-chao1,2, LIU Zheng-jie1,2( )
)
Received:2024-06-21
Online:2025-01-26
Published:2025-01-22
Contact:
LIU Zheng-jie
E-mail:2652576980@qq.com;2013045@ynau.edu.cn
LI Yun-bin, LI Yu-ping, CHENG Qin, LIN Chun, MAO Zi-chao, LIU Zheng-jie. Identification and Expression Analysis of the Members of the Asparagus officinalis DUF247 Gene Family[J]. Biotechnology Bulletin, 2025, 41(1): 198-209.

Fig. 1 Phylogenetic tree of DUF247 protein from A. officinalis, A. setaceus, A. taliensis and A. thaliana V1: A. officinalis; OF: A. setaceus; Ata: A. taliensis; AT: A. thaliana
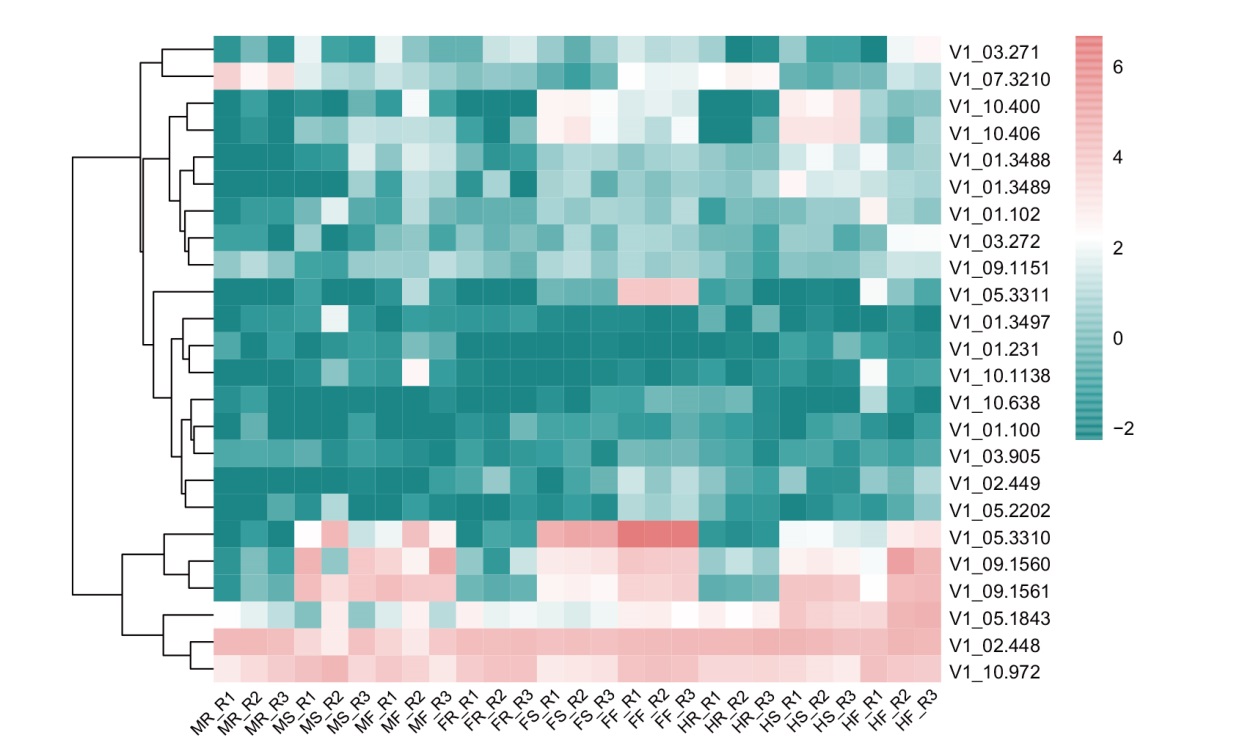
Fig. 4 Heatmaps of expressions of gene members of A. officialis DUF247 family in different genders and tissues or organs MR: Male root; MS: male stem; MF: male flower; FR: female root; FS: female stem; FF: female flower; HR: hermaphrodite root; HS: hermaphrodite stem; HF: hermaphrodite flower; R1, R2, and R3 indicate the three replicates of the sample. The same below
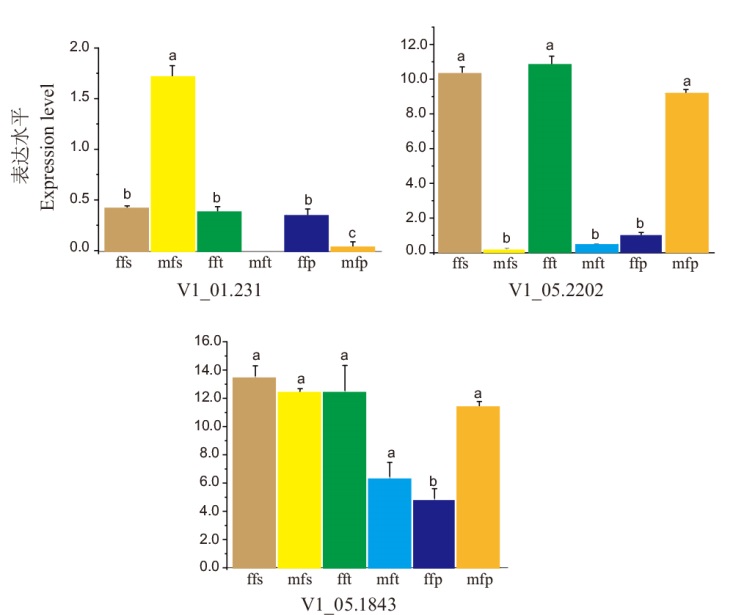
Fig. 5 Different expressions of V1_01.231, V1_05.1843, and V1_05.2202 in early development buds of different genders ffs: Female flower'stamen; mfs: male flower'stamen; fft: female flower'tapal; mft: male flower'tapal; ffp: female flower'pistil; mfp: male flower'pistil. Different lowercase letters indicatet significant differences(P<0.05), the same below
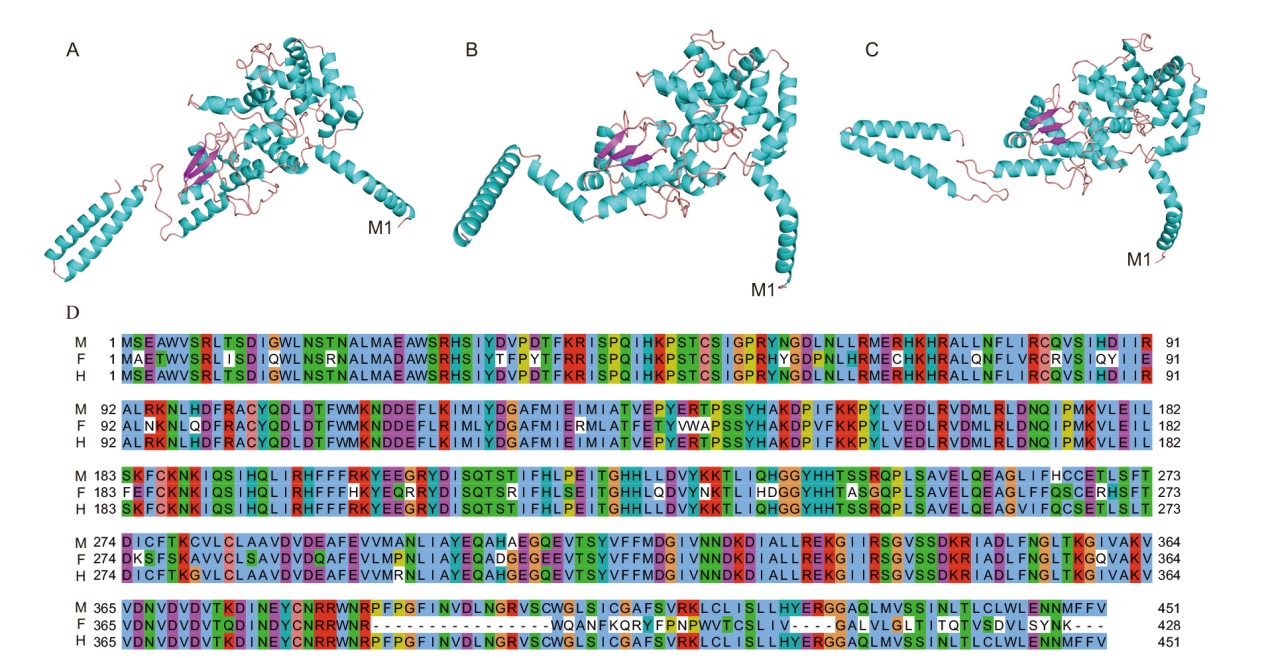
Fig. 6 Primary structure comparison and 3D structure prediction of protein encoded by AoSOFF gene in A. officinalis A: 3D structure of AoSOFF-M protein; B: 3D structure of AoSOFF-F protein; C: 3D structure of AoSOFF-H protein; D: comparison of three proteins in primary structure, M for male, F for female, and H for hermaphroditic
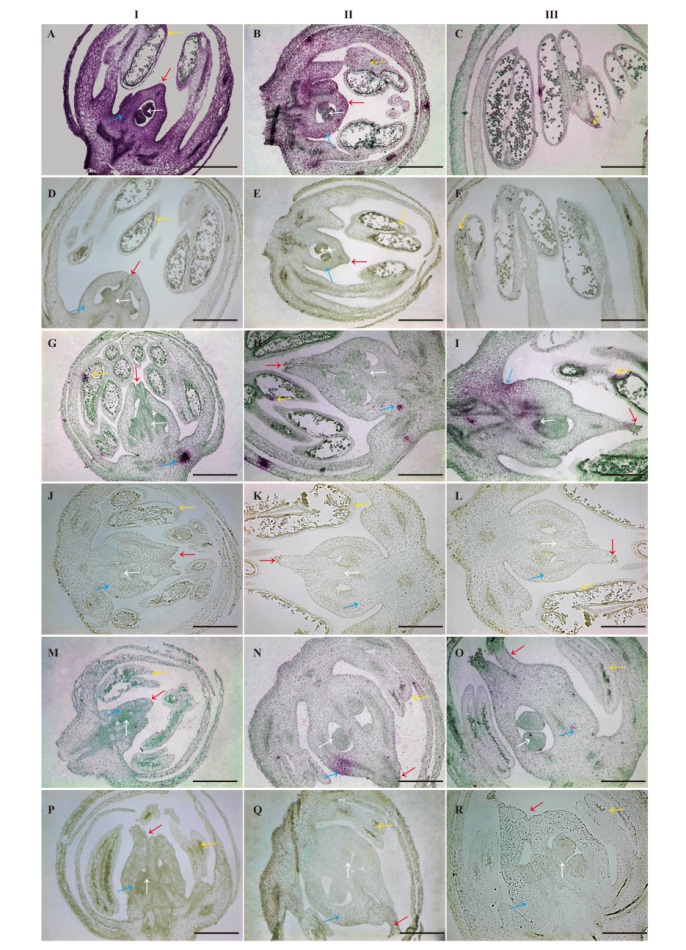
Fig. 8 Detection of A. officinalis AoSOFF in male, female and hermaphroditic flowers by in situ hybridization A-C: Male flower; D-F: male flower control; G-I: hermaphroditic flower; J-L: hermaphroditic flower control; M-O: female flower in situ hybridization results; P-R: female flower control. I, II, and III: Three stages of flower development. White arrow: Ovule; blue arrow: ovary; red arrows: stigma; yellow arrow: outer wall of pollen sac. Bar=500 μm
| [1] | Mistry J, Chuguransky S, Williams L, et al. Pfam: the protein families database in 2021[J]. Nucleic Acids Res, 2021, 49(D1): D412-D419. |
| [2] | Temple H, Mortimer JC, Tryfona T, et al. Two members of the DUF579 family are responsible for arabinogalactan methylation in Arabidopsis[J]. Plant Direct, 2019, 3(2): e00117. |
| [3] | Li XJ, Sun LJ, Tan LB, et al. TH1, a DUF640 domain-like gene controls lemma and palea development in rice[J]. Plant Mol Biol, 2012, 78(4/5): 351-359. |
| [4] | Yan DW, Zhou Y, Ye SH, et al. BEAK-SHAPED GRAIN 1/TRIANGULAR HULL 1, a DUF640 gene, is associated with grain shape, size and weight in rice[J]. Sci China Life Sci, 2013, 56(3): 275-283. |
| [5] |
罗璇, 林正雨, 陈章, 等. 玉米DUF1685基因家族的鉴定和进化分析[J]. 热带作物学报, 2022, 43(12): 2431-2442.
doi: 10.3969/j.issn.1000-2561.2022.12.005 |
| Luo X, Lin ZY, Chen Z, et al. Identification and evolutionary analysis of maize DUF1685 gene family[J]. Chin J Trop Crops, 2022, 43(12): 2431-2442. | |
| [6] |
Shinozuka H, Cogan NOI, Smith KF, et al. Fine-scale comparative genetic and physical mapping supports map-based cloning strategies for the self-incompatibility loci of perennial ryegrass(Lolium perenne L.)[J]. Plant Mol Biol, 2010, 72(3): 343-355.
doi: 10.1007/s11103-009-9574-y pmid: 19943086 |
| [7] | Zhang XJ, Jia Y, Liu Y, et al. Challenges and perspectives in the study of self-incompatibility in orchids[J]. Int J Mol Sci, 2021, 22(23): 12901. |
| [8] |
Liang M, Cao ZH, Zhu AD, et al. Evolution of self-compatibility by a mutant Sm-RNase in citrus[J]. Nat Plants, 2020, 6(2): 131-142.
doi: 10.1038/s41477-020-0597-3 pmid: 32055045 |
| [9] | Ma L, Zhang CZ, Zhang B, et al. A nonS-locus F-box gene breaks self-incompatibility in diploid potatoes[J]. Nat Commun, 2021, 12(1): 4142. |
| [10] | Cheung AY. Self-incompatibility in Papaver rhoeas: a role for ATP[J]. New Phytol, 2022, 236(5): 1625-1628. |
| [11] |
Gervais C, Awad DA, Roze D, et al. Genetic architecture of inbreeding depression and the maintenance of gametophytic self-incompatibility[J]. Evolution, 2014, 68(11): 3317-3324.
doi: 10.1111/evo.12495 pmid: 25065256 |
| [12] | Li C, Long Y, Lu MQ, et al. Gene coexpression analysis reveals key pathways and hub genes related to late-acting self-incompatibility in Camellia oleifera[J]. Front Plant Sci, 2023, 13: 1065872. |
| [13] |
Silva NF, Goring DR. Mechanisms of self-incompatibility in flowering plants[J]. Cell Mol Life Sci, 2001, 58(14): 1988-2007.
doi: 10.1007/PL00000832 pmid: 11814052 |
| [14] |
Hiscock SJ, McInnis SM. Pollen recognition and rejection during the sporophytic self-incompatibility response: Brassica and beyond[J]. Trends Plant Sci, 2003, 8(12): 606-613.
pmid: 14659710 |
| [15] | Herridge R, McCourt T, Jacobs JME, et al. Identification of the genes at S and Z reveals the molecular basis and evolution of grass self-incompatibility[J]. Front Plant Sci, 2022, 13: 1011299. |
| [16] | Lian XP, Zhang SL, Huang GF, et al. Confirmation of a gametophytic self-incompatibility in Oryza longistaminata[J]. Front Plant Sci, 2021, 12: 576340. |
| [17] |
Harkess A, Zhou JS, Xu CY, et al. The asparagus genome sheds light on the origin and evolution of a young Y chromosome[J]. Nat Commun, 2017, 8(1): 1279.
doi: 10.1038/s41467-017-01064-8 pmid: 29093472 |
| [18] | 刘江, 刘刚, 曾益春, 等. 桑树GATA基因家族的全基因组鉴定与分析[J]. 西南农业学报, 2022, 35(12): 2724-2731. |
| Liu J, Liu G, Zeng YC, et al. Genome-wide identification and analysis of mulberry(Morus notabilis)GATA gene family[J]. Southwest China J Agric Sci, 2022, 35(12): 2724-2731. | |
| [19] | 陈曙, 张彧, 陈卓, 等. 玉米泛素连接酶U-box基因家族的全基因组鉴定及表达分析[J]. 西南农业学报, 2022, 35(3): 481-490. |
| Chen S, Zhang Y, Chen Z, et al. Genome-wide identification and expression analysis of ubiquitin ligase U-box gene family in maize[J]. Southwest China J Agric Sci, 2022, 35(3): 481-490. | |
| [20] |
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput[J]. Nucleic Acids Res, 2004, 32(5): 1792-1797.
doi: 10.1093/nar/gkh340 pmid: 15034147 |
| [21] |
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies[J]. Bioinformatics, 2014, 30(9): 1312-1313.
doi: 10.1093/bioinformatics/btu033 pmid: 24451623 |
| [22] |
Letunic I, Bork P. Interactive Tree Of Life(iTOL): an online tool for phylogenetic tree display and annotation[J]. Bioinformatics, 2007, 23(1): 127-128.
doi: 10.1093/bioinformatics/btl529 pmid: 17050570 |
| [23] | Bailey TL, Williams N, Misleh C, et al. MEME: discovering and analyzing DNA and protein sequence motifs[J]. Nucleic Acids Res, 2006, 34(Web Server issue): W369-W373. |
| [24] | Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server[J]. Bioinformatics, 2015, 31(8): 1296-1297. |
| [25] |
Wang YP, Li JP, Paterson AH. MCScanX-transposed: detecting transposed gene duplications based on multiple colinearity scans[J]. Bioinformatics, 2013, 29(11): 1458-1460.
doi: 10.1093/bioinformatics/btt150 pmid: 23539305 |
| [26] | Wen GZ. A simple process of RNA-sequence analyses by Hisat2, htseq and DESeq2[C]// Proceedings of the 2017 International Conference on Biomedical Engineering and Bioinformatics. Bangkok Thailand. New York: ACM, 2017: 11. |
| [27] |
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features[J]. Bioinformatics, 2014, 30(7): 923-930.
doi: 10.1093/bioinformatics/btt656 pmid: 24227677 |
| [28] |
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2009, 25(14): 1754-1760.
doi: 10.1093/bioinformatics/btp324 pmid: 19451168 |
| [29] |
Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer[J]. Nat Biotechnol, 2011, 29(1): 24-26.
doi: 10.1038/nbt.1754 pmid: 21221095 |
| [30] | Delano W L. Pymol: An open-source molecular graphics tool[J]. Ccp4 Newslett Protein Crystallogr, 2002, 40(1): 82-92. |
| [31] |
Waterhouse AM, Procter JB, Martin DMA, et al. Jalview Version 2—a multiple sequence alignment editor and analysis workbench[J]. Bioinformatics, 2009, 25(9): 1189-1191.
doi: 10.1093/bioinformatics/btp033 pmid: 19151095 |
| [32] | 谷倩楠, 孔瑞文, 孙明哲, 等. 大豆混合盐碱胁迫应答基因GmDUF247-1的克隆及功能分析[J]. 植物遗传资源学报, 2024, 25(6): 978-989. |
| Gu QN, Kong RW, Sun MZ, et al. Cloning and functional characterization of the GmDUF247-1 gene in soybean response to saline-alkaline stress[J]. J Plant Genet Resour, 2024, 25(06): 978-989. | |
| [33] | Zhang FF, Liu YX, Liu F, et al. Genome-wide characterization and analysis of rice DUF247 gene family[J]. BMC Genomics, 2024, 25(1): 613. |
| [34] |
Manzanares C, Barth S, Thorogood D, et al. A gene encoding a DUF247 domain protein cosegregates with the S self-incompatibility locus in perennial ryegrass[J]. Mol Biol Evol, 2016, 33(4): 870-884.
doi: 10.1093/molbev/msv335 pmid: 26659250 |
| [35] | Shin NR, Shin YH, Kim HS, et al. Function analysis of the PR55/B gene related to self-incompatibility in Chinese cabbage using CRISPR/Cas9[J]. Int J Mol Sci, 2022, 23(9): 5062. |
| [36] | Sun LH, Cao SY, Zheng N, et al. Analyses of Cullin1 homologs reveal functional redundancy in S-RNase-based self-incompatibility and evolutionary relationships in eudicots[J]. Plant Cell, 2023, 35(2): 673-699. |
| [1] | XU Xue-fei, YANG Pan-pan, ZHANG Wen-liang, BIAN Guang-ya, XU Lei-feng, LIU Hui-chao, MING Jun. Distribution of Lily Mottle Virus in the Shoot Tips and Root Tips of Lilium lancifolium [J]. Biotechnology Bulletin, 2024, 40(8): 212-220. |
| [2] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [3] | LI Xin-yi, JIANG Chun-xiu, XUE Li, JIANG Hong-tao, YAO Wei, DENG Zu-hu, ZHANG Mu-qing, YU Fan. Enhancing Hybridization Signal of Sugarcane Chromosome Oligonucleotide Probe via Multiple Fluorescence Labeled Primers [J]. Biotechnology Bulletin, 2023, 39(5): 103-111. |
| [4] | CAI Meng-xian, GAO Zuo-min, HU Li-juan, FENG Qun, WANG Hong-cheng, ZHU Bin. Development and Genetic Analysis of Two Nullisomic Lines(NC1 and NC2)in Natural Brassica napus [J]. Biotechnology Bulletin, 2023, 39(3): 81-88. |
| [5] | GAO Li-li, DIAO Xiao-ming, LI Yun, ZHAI Xu-liang, ZHOU Chun-long. Advances and Reflect on Dominant Factors Determining the Sex of Pelodiscus sinensis [J]. Biotechnology Bulletin, 2018, 34(12): 41-49. |
| [6] | Dong Tian, Liu Fengjiao, Hu Hongxia, Zhu Hua. Research Progress on Sex Differentiation Related Genes on Sturgeon [J]. Biotechnology Bulletin, 2015, 31(10): 62-70. |
| [7] | Wang Risheng, Zhang Man, Huang Rukui, Liu Wenjun, Dong Wenbin, Che Jianglü, Fang Fengxue, Li Yangrui. Cloning and Sequence Analysis of 1-aminocyclopropane-1-carboxylate (ACC)Oxidase Gene cDNA from Gynoecious Momordica charantia [J]. Biotechnology Bulletin, 2013, 29(10): 76-80. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||