








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (1): 210-220.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0507
Previous Articles Next Articles
KONG Qing-yang( ), ZHANG Xiao-long, LI Na, ZHANG Chen-jie, ZHANG Xue-yun, YU Chao, ZHANG Qi-xiang, LUO Le(
), ZHANG Xiao-long, LI Na, ZHANG Chen-jie, ZHANG Xue-yun, YU Chao, ZHANG Qi-xiang, LUO Le( )
)
Received:2024-05-29
Online:2025-01-26
Published:2025-01-22
Contact:
LUO Le
E-mail:kongqingyangd@163.com;luolebjfu@163.com
KONG Qing-yang, ZHANG Xiao-long, LI Na, ZHANG Chen-jie, ZHANG Xue-yun, YU Chao, ZHANG Qi-xiang, LUO Le. Identification and Expression Analysis of GRAS Transcription Factor Family in Rosa persica[J]. Biotechnology Bulletin, 2025, 41(1): 210-220.

Fig. 2 Phylogeny, conserved motifs and gene structure analysis of GRAS family proteins in R. persica The scale indicates protein sequence length(aa)and gene length(bp), respectively

Fig. 4 Internal collinearity analysis of GRAS gene family of R. persica and the collinearity analysis of GRAS gene family R. persica and Arabidopsis thaliana A: Collinearity analysis of the GRAS gene family in R. persica. B: Analysis of collinearity between the GRAS gene family of R. persica and Arabidopsis thaliana
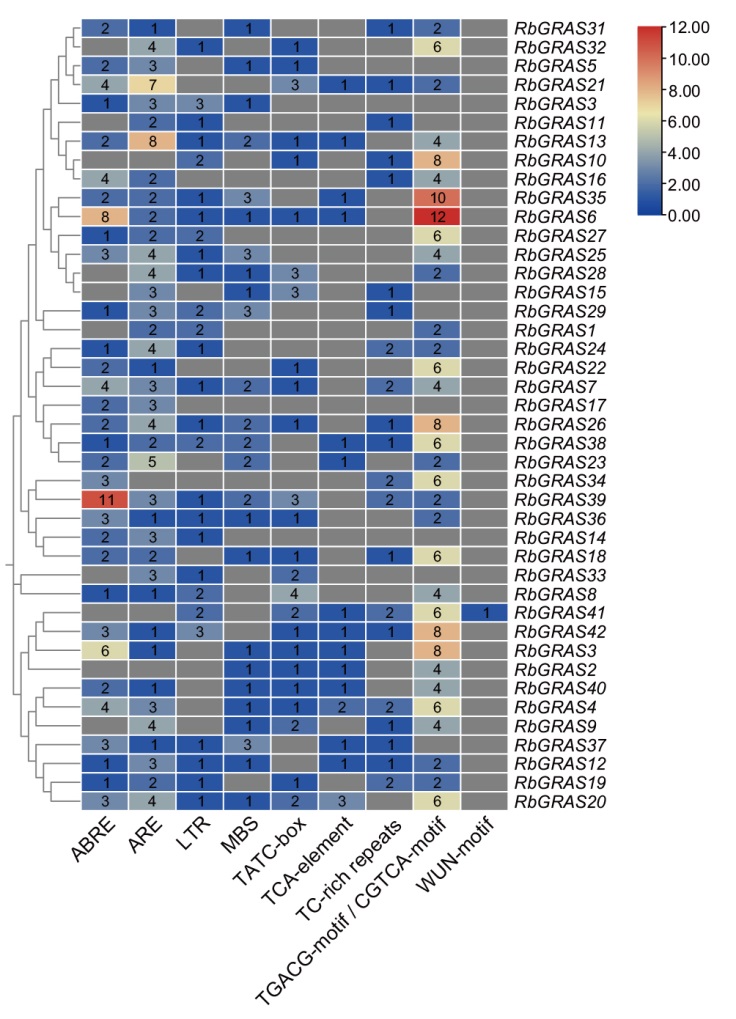
Fig. 6 Statistics on the number of cis-acting elements in the GRAS gene promoter of R. persica TC-rich repeats: Defense and stress response elements; ABRE: abscisic acid response element; ARE: anaerobic induction regulatory element; TGACG-motif/CGTCA-motif: methyl jasmonate response element; MBS: drought-induced element; TATC-box: gibberellin response element; LTR: low temperature response element; TCA-element: salicylic acid response element; WUN-motif: wound-responsive element

Fig. 8 Heatmap of GRAS gene expression in different tissues of R. persica under different stresses Root, stem, leaf, flower, fruit: Non-stressed roots, stems, leaves, flowers and fruits; CKr, CKl: control group roots and leaves; LDr, LDl: roots and leaves under mild drought stress; SDr, SDl: roots and leaves under severe drought stress. G9-G4: Leaves from September 2021 to April 2022 in the wild environment. J9-J4: Stems from September 2021 to April 2022 in the wild environment
| [1] | 新疆植物志编辑委员会. 新疆植物志-第二卷, 第二分册-小檗科—蔷薇科[M]. 乌鲁木齐: 新疆科技卫生出版社, 1995. |
| Editorial Committee for the Flora of Xinjiang. Flora xinjiangensis - Volume two, part two - Berberidaceae to Rosaceae[M]. Xinjiang Science, Technology and Public Health Press, 1995. | |
| [2] | 惠俊爱, 张霞, 王绍明. 新疆野生单叶蔷薇生物学特性分析[J]. 山东林业科技, 2013, 43(4): 61-63. |
| Hui JA, Zhang X, Wang SM. Analysis of biological characteristics of wild Hulthemia berberifolia(pall.)dumort in Xinjiang[J]. J Shandong For Sci Technol, 2013, 43(4): 61-63. | |
| [3] | 中国科学院中国植物志编辑委员会. 中国植物志-第七十四卷[M]. 北京: 科学出版社, 1985: 370-371. |
| Committee on the Flora of China, Chinese Academy of Sciences. Flora of China(37 vols)[M]. Beijing: Science Press, 1985: 370-371. | |
| [4] | 丛者福. 新疆野蔷薇果的研究[J]. 干旱区资源与环境, 1996, 10(4): 100-102. |
| Cong ZF. Study on the fruit of wild rose in Xinjiang[J]. Arid Land Resour Environ, 1996, 10(4): 100-102. | |
| [5] | 范春国. 单叶蔷薇耐旱关键转录因子挖掘与功能初步验证[D]. 南京: 南京农业大学, 2021. |
| Fan CG. Mining and functional verification of key transcription factors of drought tolerance in Rosa persica[D]. Nanjing: Nanjing Agricultural University, 2021. | |
| [6] |
冯策婷, 江律, 刘鑫颖, 等. 单叶蔷薇NAC基因家族鉴定及干旱胁迫响应分析[J]. 生物技术通报, 2023, 39(11): 283-296.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0531 |
| Feng CT, Jiang L, Liu XY, et al. Identification of the NAC gene family in Rosa persica and response analysis under drought stress[J]. Biotechnol Bull, 2023, 39(11): 283-296. | |
| [7] | Zhuang YY, Zhou LJ, Geng LF, et al. Genome-wide identification of the bHLH transcription factor family in Rosa persica and response to low-temperature stress[J]. PeerJ, 2024, 12: e16568. |
| [8] | Moradkhani S, Rezaei-Dehghanzadeh T, Nili-Ahmadabadi A. Rosa persica hydroalcoholic extract improves cadmium-hepatotoxicity by modulating oxidative damage and tumor necrosis factor-alpha status[J]. Environ Sci Pollut Res Int, 2020, 27(25): 31259-31268. |
| [9] | 欧哲, 杨宇, 冯策婷, 等. 单叶蔷薇远缘杂交中花粉管生长的荧光显微观察[J]. 东北农业大学学报, 2022, 53(10): 18-26. |
| Ou Z, Yang Y, Feng CT, et al. Fluorescent microscope observation on growth of pollen tube on distant hybridization in Rosa persica[J]. J Northeast Agric Univ, 2022, 53(10): 18-26. | |
| [10] |
Pysh LD, Wysocka-Diller JW, Camilleri C, et al. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes[J]. Plant J, 1999, 18(1): 111-119.
pmid: 10341448 |
| [11] |
Sun XL, Xue B, Jones WT, et al. A functionally required unfoldome from the plant Kingdom: intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development[J]. Plant Mol Biol, 2011, 77(3): 205-223.
doi: 10.1007/s11103-011-9803-z pmid: 21732203 |
| [12] | Khan Y, Xiong Z, Zhang H, et al. Expression and roles of GRAS gene family in plant growth, signal transduction, biotic and abiotic stress resistance and symbiosis formation-a review[J]. Plant Biol, 2022, 24(3): 404-416. |
| [13] | Tian CG, Wan P, Sun SH, et al. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis[J]. Plant Mol Biol, 2004, 54(4): 519-532. |
| [14] |
Li Z, Tu QC, Lyu XG, et al. GmSTF accumulation mediated by DELLA protein GmRGAs contributes to coordinating light and gibberellin signaling to reduce plant height in soybean[J]. Crop J, 2024, 12(2): 432-442.
doi: 10.1016/j.cj.2024.01.013 |
| [15] |
Helariutta Y, Fukaki H, Wysocka-Diller J, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling[J]. Cell, 2000, 101(5): 555-567.
doi: 10.1016/s0092-8674(00)80865-x pmid: 10850497 |
| [16] |
Wang ZM, Wong DCJ, Wang Y, et al. GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response[J]. Plant Physiol, 2021, 186(3): 1660-1678.
doi: 10.1093/plphys/kiab142 pmid: 33752238 |
| [17] | Zhang S, Li XW, Fan SD, et al. Overexpression of HcSCL13, a Halostachys caspica GRAS transcription factor, enhances plant growth and salt stress tolerance in transgenic Arabidopsis[J]. Plant Physiol Biochem, 2020, 151: 243-254. |
| [18] |
Silverstone AL, Ciampaglio CN, Sun T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway[J]. Plant Cell, 1998, 10(2): 155-169.
doi: 10.1105/tpc.10.2.155 pmid: 9490740 |
| [19] |
Dinneny JR, Benfey PN. Plant stem cell niches: standing the test of time[J]. Cell, 2008, 132(4): 553-557.
doi: 10.1016/j.cell.2008.02.001 pmid: 18295573 |
| [20] |
Han H, Yan A, Li LH, et al. A signal cascade originated from epidermis defines apical-basal patterning of Arabidopsis shoot apical meristems[J]. Nat Commun, 2020, 11(1): 1214.
doi: 10.1038/s41467-020-14989-4 pmid: 32139673 |
| [21] |
Morohashi K, Minami M, Takase H, et al. Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression[J]. J Biol Chem, 2003, 278(23): 20865-20873.
doi: 10.1074/jbc.M301712200 pmid: 12657631 |
| [22] | 孙彦琳, 于超, 罗乐. 单叶蔷薇bZIP转录因子家族鉴定与表达分析[J]. 西北农林科技大学学报: 自然科学版, 2022, 50(6): 82-92. |
| Sun YL, Yu C, Luo L, et al. Identification and expression analysis of bZIP transcription factor family in Rosa persica[J]. J Northwest A F Univ Nat Sci Ed, 2022, 50(6): 82-92. | |
| [23] |
于婷婷, 李欢, 宁源生, 等. 苹果GRAS全基因组鉴定及其对生长素的响应分析[J]. 园艺学报, 2023, 50(2): 397-409.
doi: 10.16420/j.issn.0513-353x.2021-1039 |
| Yu TT, Li H, Ning YS, et al. Genome-wide identification of GRAS gene family in apple and expression analysis of its response to auxin[J]. Acta Hortic Sin, 2023, 50(2): 397-409. | |
| [24] | Kumari P, Gahlaut V, Kaur E, et al. Genome-wide identification of GRAS transcription factors and their potential roles in growth and development of rose(Rosa chinensis)[J]. J Plant Growth Regul, 2023, 42(3): 1505-1521. |
| [25] | 牛义岭. 番茄GRAS基因家族生物信息学分析及部分抗性相关基因鉴定分析[D]. 哈尔滨: 东北农业大学, 2017. |
| Niu YL. Bioinformatics analysis of GRAS gene family in tomato and identification of some resistance-related genes[D]. Harbin: Northeast Agricultural University, 2017. | |
| [26] | 惠俊爱, 张霞, 王绍明. 新疆野生单叶蔷薇的染色体核型分析[J]. 山东林业科技, 2013, 43(4): 58-60. |
| Hui JA, Zhang X, Wang SM. Karyotype analysis of Xin Jiang wild Hulthemia berberifolia(Pall.)Dumort[J]. J Shandong For Sci Technol, 2013, 43(4): 58-60. | |
| [27] |
Shaul O. How introns enhance gene expression[J]. Int J Biochem Cell Biol, 2017, 91(Pt B): 145-155.
doi: S1357-2725(17)30154-1 pmid: 28673892 |
| [28] | Cannon SB, Mitra A, Baumgarten A, et al. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana[J]. BMC Plant Biol, 2004, 4: 10. |
| [29] | Wei XP, Liu LY, Liu G, et al. Methyl jasmonate promotes suberin biosynthesis by stimulating transcriptional activation of AchMYC2 on AchFHT in wound healing of kiwifruit[J]. Postharvest Biol Technol, 2024, 210: 112741. |
| [30] | 赵曼如, 胡文忠, 于皎雪, 等. 茉莉酸甲酯对果蔬抗性、抗氧化活性及品质影响的研究进展[J]. 食品工业科技, 2020, 41(4): 328-332. |
| Zhao MR, Hu WZ, Yu JX, et al. Research progress on effects of methyl jasmonate on resistance, antioxidant activity and quality of fruits and vegetables[J]. Sci Technol Food Ind, 2020, 41(4): 328-332. | |
| [31] | Feng M, Zhang A, Nguyen V, et al. A conserved graft formation process in Norway spruce and Arabidopsis identifies the PAT gene family as central regulators of wound healing[J]. Nat Plants, 2024, 10(1): 53-65. |
| [32] |
Rivas-San Vicente M, Plasencia J. Salicylic acid beyond defence: its role in plant growth and development[J]. J Exp Bot, 2011, 62(10): 3321-3338.
doi: 10.1093/jxb/err031 pmid: 21357767 |
| [33] | 龙亚芹, 王万东, 王美存, 等. 水杨酸(SA)诱导植物对病虫害产生抗性及作用机制研究[J]. 热带农业科学, 2009, 29(12): 46-50. |
| Long YQ, Wang WD, Wang MC, et al. Salicylic acid induced resistance of plants against insects and diseases and its interaction mechanism[J]. Chin J Trop Agric, 2009, 29(12): 46-50. | |
| [34] | Incarbone M, Bradamante G, Pruckner F, et al. Salicylic acid and RNA interference mediate antiviral immunity of plant stem cells[J]. Proc Natl Acad Sci U S A, 2023, 120(42): e2302069120. |
| [35] | Li SL, Li XN, Wei ZH, et al. ABA-mediated modulation of elevated CO2 on stomatal response to drought[J]. Curr Opin Plant Biol, 2020, 56: 174-180. |
| [36] | Zhong MS, Jiang H, Cao Y, et al. MdCER2 conferred to wax accumulation and increased drought tolerance in plants[J]. Plant Physiol Biochem, 2020, 149: 277-285. |
| [37] | 张晓龙, 邓童, 刘学森, 等. 单叶蔷薇幼苗根系对不同潜水埋深的适应机制[J]. 生态学报, 2022, 42(15): 6137-6149. |
| Zhang XL, Deng T, Liu XS, et al. Adaptability mechanism of Rosa persica seedlings root in different groundwater levels[J]. Acta Ecol Sin, 2022, 42(15): 6137-6149. | |
| [38] | Lucas M, Swarup R, Paponov IA, et al. Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis[J]. Plant Physiol, 2011, 155(1): 384-398. |
| [1] | LI Yu-xin, LI Miao, DU Xiao-fen, HAN Kang-ni, LIAN Shi-chao, WANG Jun. Identification and Expression Analysis of SiSAP Gene Family in Foxtail Millet(Setaria italica) [J]. Biotechnology Bulletin, 2025, 41(1): 143-156. |
| [2] | WANG Zi-ao, TIAN Rui, CUI Yong-mei, BAI Yi-xiong, YAO Xiao-hua, AN Li-kun, WU Kun-lun. Bioinformatics and Expression Pattern Analysis of HvnJAZ4 Gene in Hulless Barley [J]. Biotechnology Bulletin, 2025, 41(1): 173-185. |
| [3] | SONG Bing-fang, LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi. Identification of Potato G6PDH Gene Family and Its Expression Analysis in Damaged Tubers [J]. Biotechnology Bulletin, 2024, 40(9): 104-112. |
| [4] | WU Hui-qin, WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan. Identification and Expression Analysis of UGT Gene Family in Pepper [J]. Biotechnology Bulletin, 2024, 40(9): 198-211. |
| [5] | MAN Quan-cai, MENG Zi-nuo, LI Wei, CAI Xin-ru, SU Run-dong, FU Chang-qing, GAO Shun-juan, CUI Jiang-hui. Identification and Expression Analysis of AQP Gene Family in Potato [J]. Biotechnology Bulletin, 2024, 40(9): 51-63. |
| [6] | WU Juan, WU Xiao-juan, WANG Pei-jie, XIE Rui, NIE Hu-shuai, LI Nan, MA Yan-hong. Screening and Expression Analysis of ERF Gene Related to Anthocyanin Synthesis in Colored Potato [J]. Biotechnology Bulletin, 2024, 40(9): 82-91. |
| [7] | SHEN Peng, GAO Ya-Bin, DING Hong. Identification and Expression Analysis of SAT Gene Family in Potato(Solanum tuberosum L.) [J]. Biotechnology Bulletin, 2024, 40(9): 64-73. |
| [8] | LI Yi-jun, YANG Xiao-bei, XIA Lin, LUO Zhao-peng, XU Xin, YANG Jun, NING Qian-ji, WU Ming-zhu. Cloning and Functional Analysis of NtPRR37 Gene in Nicotiana tabacum L. [J]. Biotechnology Bulletin, 2024, 40(8): 221-231. |
| [9] | ZHOU Ran, WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma. Analysis of LncRNA Differential Expression in Mammary Tissue of Cows with Staphylococcus aureus Mastitis [J]. Biotechnology Bulletin, 2024, 40(8): 320-328. |
| [10] | WU Shuai, XIN Yan-ni, MAI Chun-hai, MU Xiao-ya, WANG Min, YUE Ai-qin, ZHAO Jin-zhong, WU Shen-jie, DU Wei-jun, WANG Li-xiang. Genome-wide Identification and Stress Response Analysis of Soybean GS Gene Family [J]. Biotechnology Bulletin, 2024, 40(8): 63-73. |
| [11] | CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage [J]. Biotechnology Bulletin, 2024, 40(8): 74-82. |
| [12] | HAN Kai, ZHOU Yong-shun, ZHANG Kai-yue, WANG Lu, GAO Jian-feng, CHEN Fu-long. Evaluation of Drought Resistance of Three Chlorella Strains [J]. Biotechnology Bulletin, 2024, 40(8): 244-254. |
| [13] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [14] | LI Yu-qing, WU Nan, LUO Jian-rang. Cloning and Functional Analysis of bHLH Gene Related to Anthocyanin Synthesis in Paeonia qiui [J]. Biotechnology Bulletin, 2024, 40(8): 174-185. |
| [15] | ZHANG Ming-ya, PANG Sheng-qun, LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong. Identification and Expression Analysis of FAD Gene Family in Solanum lycopersicum [J]. Biotechnology Bulletin, 2024, 40(7): 150-162. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||