








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (4): 243-255.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0883
HUANG Jin-heng1( ), HUANG Xi1, ZHANG Jia-yan1, ZHOU Xin-yu1, LIAO Pei-ran1,2(
), HUANG Xi1, ZHANG Jia-yan1, ZHOU Xin-yu1, LIAO Pei-ran1,2( ), YANG Quan1,2,3(
), YANG Quan1,2,3( )
)
Received:2024-09-11
Online:2025-04-26
Published:2025-04-25
Contact:
LIAO Pei-ran, YANG Quan
E-mail:2687881471@qq.com;westpp@126.com;yangquan7208@vip.163.com
HUANG Jin-heng, HUANG Xi, ZHANG Jia-yan, ZHOU Xin-yu, LIAO Pei-ran, YANG Quan. Identification and Expression Analysis of the C3H Gene Family in Grona styracifolia across Different Varieties[J]. Biotechnology Bulletin, 2025, 41(4): 243-255.
正向引物名称 Forward primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物名称 Reverse primer name | 反向引物序列 Reverse primer sequence (5′‒3′) |
|---|---|---|---|
| GsC3H1f | AACCAGAACCATTGGAGGAAA | GsC3H1r | TGATAGGCGACAGTTACTACCAAAT |
| GsC3H2f | CTTCCCTTGGAGCATACCCT | GsC3H2r | GAGGCACATAAGATTGAAGACCA |
| GsC3H3f | GCACTGTCTCCATCTCCGTTAC | GsC3H3r | TTGACCATCTGCTTTGACCC |
| GsC3H4f | GTCAAATGCCGTCAACAGGA | GsC3H4r | CGCAGATAGCGAAGGAAGGT |
| GsC3H5f | CGATTCCGCAGCACTTTCC | GsC3H5r | CACCCTTACCAGTTTCTTCGTCACA |
| GsC3H6f | CAACGCAGGTATGGAATCAAA | GsC3H6r | GGTAGACCCTTGTCACTGAGGTT |
| GsC3H7f | ATGGTGTAGTTACCTTATGTGGGATG | GsC3H7r | TGCTCGGACCTCTTGTTTCG |
| GsC3H8f | TACAAAGCGGAGCCAGAGC | GsC3H8r | GGTGGTAGCAGGTGTTAGAGGAGT |
| GsC3H9f | TCAAAGTCCGACCTTGTTCACG | GsC3H9r | TGCAGCCAGCACTCGAAAA |
| GsC3H10f | CCCCTTTGTTCACCCTGGAG | GsC3H10r | CACAGCTTGGTCCGATACTGG |
| GsC3H11f | GCTCAACTTACTAACCTGTCAGCCTC | GsC3H11r | CCGAAAGGCAGTCTTAGAGTCATC |
| GsC3H12f | GGATCTCCCAGCAAGATTGAC | GsC3H12r | GCTGCCGCTCTTTGAGTATCAT |
| GsC3H13f | CCCTCAAGATTCACTGTGGATG | GsC3H13r | TCTTTCTGGGAACTCGCCTAT |
| GsC3H14f | AGGCTGGGATTGCTGGAA | GsC3H14r | GAGATTGGACGGTAGGATAAACA |
| GsC3H15f | AAACGAAACGGACGCTGAC | GsC3H15r | GCATCGCCGAACTTTGAAC |
| GsC3H16f | TTCTCGCTCTGATCCGTCG | GsC3H16r | GCTCCAACAACCGTTCCAC |
| GsC3H17f | GCAAAGCGAAGCCAAAGTT | GsC3H17r | CTGAAAGGGCAGAGGACACC |
| GsC3H18f | CAATAGGTCGTCTTGGTGGTCG | GsC3H18r | CGCTGATCTTTGCCGTAGCA |
| GsC3H19f | CGAACACGCCGTCAACCTT | GsC3H19r | CCAAATGGGCAGCCAGAAG |
| GsC3H20f | TGAGTCTTACCCTGAGCGTCC | GsC3H20r | TGTAGCTCTTACTGCCGCATC |
| GsC3H21f | GAAGAGGAGCAGGGTAAGAAGC | GsC3H21r | CTGGCAAGTTGTGAGCAAGAGGT |
| GsC3H22f | GTCGTTTCAACCACCCTCG | GsC3H22r | TTTCTCACCCACTCGCAATG |
| GsC3H23f | ACAATGGTGACTCCGAGACAAC | GsC3H23r | GCATGTACCCGCCCTGAAT |
| GsC3H24f | AGGTGCTCCTTGTGACATTGTT | GsC3H24r | TGATTGAAAGCTCATCCCAGA |
| GsC3H25f | GGACGAAGATTTGTTGAAGCG | GsC3H25r | TCCTTCCAACGGAGGGTGT |
| GsC3H26f | TTTCTCCGATTCGCCACC | GsC3H26r | CGAGTTTAACATTCCGCATTG |
| GsC3H27f | CACCGAGCAAATCCAACCG | GsC3H27r | CACAGGAATAAGCATCGACCG |
| EF-1αf | GAGATGCCTGGTCTATTGTGGG | EF-1αr | GATTGGCTCAACTGTCAGATGCT |
Table 1 Quantitative primer sequence information
正向引物名称 Forward primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物名称 Reverse primer name | 反向引物序列 Reverse primer sequence (5′‒3′) |
|---|---|---|---|
| GsC3H1f | AACCAGAACCATTGGAGGAAA | GsC3H1r | TGATAGGCGACAGTTACTACCAAAT |
| GsC3H2f | CTTCCCTTGGAGCATACCCT | GsC3H2r | GAGGCACATAAGATTGAAGACCA |
| GsC3H3f | GCACTGTCTCCATCTCCGTTAC | GsC3H3r | TTGACCATCTGCTTTGACCC |
| GsC3H4f | GTCAAATGCCGTCAACAGGA | GsC3H4r | CGCAGATAGCGAAGGAAGGT |
| GsC3H5f | CGATTCCGCAGCACTTTCC | GsC3H5r | CACCCTTACCAGTTTCTTCGTCACA |
| GsC3H6f | CAACGCAGGTATGGAATCAAA | GsC3H6r | GGTAGACCCTTGTCACTGAGGTT |
| GsC3H7f | ATGGTGTAGTTACCTTATGTGGGATG | GsC3H7r | TGCTCGGACCTCTTGTTTCG |
| GsC3H8f | TACAAAGCGGAGCCAGAGC | GsC3H8r | GGTGGTAGCAGGTGTTAGAGGAGT |
| GsC3H9f | TCAAAGTCCGACCTTGTTCACG | GsC3H9r | TGCAGCCAGCACTCGAAAA |
| GsC3H10f | CCCCTTTGTTCACCCTGGAG | GsC3H10r | CACAGCTTGGTCCGATACTGG |
| GsC3H11f | GCTCAACTTACTAACCTGTCAGCCTC | GsC3H11r | CCGAAAGGCAGTCTTAGAGTCATC |
| GsC3H12f | GGATCTCCCAGCAAGATTGAC | GsC3H12r | GCTGCCGCTCTTTGAGTATCAT |
| GsC3H13f | CCCTCAAGATTCACTGTGGATG | GsC3H13r | TCTTTCTGGGAACTCGCCTAT |
| GsC3H14f | AGGCTGGGATTGCTGGAA | GsC3H14r | GAGATTGGACGGTAGGATAAACA |
| GsC3H15f | AAACGAAACGGACGCTGAC | GsC3H15r | GCATCGCCGAACTTTGAAC |
| GsC3H16f | TTCTCGCTCTGATCCGTCG | GsC3H16r | GCTCCAACAACCGTTCCAC |
| GsC3H17f | GCAAAGCGAAGCCAAAGTT | GsC3H17r | CTGAAAGGGCAGAGGACACC |
| GsC3H18f | CAATAGGTCGTCTTGGTGGTCG | GsC3H18r | CGCTGATCTTTGCCGTAGCA |
| GsC3H19f | CGAACACGCCGTCAACCTT | GsC3H19r | CCAAATGGGCAGCCAGAAG |
| GsC3H20f | TGAGTCTTACCCTGAGCGTCC | GsC3H20r | TGTAGCTCTTACTGCCGCATC |
| GsC3H21f | GAAGAGGAGCAGGGTAAGAAGC | GsC3H21r | CTGGCAAGTTGTGAGCAAGAGGT |
| GsC3H22f | GTCGTTTCAACCACCCTCG | GsC3H22r | TTTCTCACCCACTCGCAATG |
| GsC3H23f | ACAATGGTGACTCCGAGACAAC | GsC3H23r | GCATGTACCCGCCCTGAAT |
| GsC3H24f | AGGTGCTCCTTGTGACATTGTT | GsC3H24r | TGATTGAAAGCTCATCCCAGA |
| GsC3H25f | GGACGAAGATTTGTTGAAGCG | GsC3H25r | TCCTTCCAACGGAGGGTGT |
| GsC3H26f | TTTCTCCGATTCGCCACC | GsC3H26r | CGAGTTTAACATTCCGCATTG |
| GsC3H27f | CACCGAGCAAATCCAACCG | GsC3H27r | CACAGGAATAAGCATCGACCG |
| EF-1αf | GAGATGCCTGGTCTATTGTGGG | EF-1αr | GATTGGCTCAACTGTCAGATGCT |
基因名 Gene name | 氨基酸数量 Amino acid count/aa | 相对分子质量 Relative molecular mass/Da | 等电点 Isoelectric point | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 亲水性 Hydrophilicity |
|---|---|---|---|---|---|---|
| GsC3H1 | 495 | 55 557.14 | 5.54 | 49.44 | 70.95 | -0.761 |
| GsC3H2 | 424 | 46 321.03 | 8.10 | 58.80 | 54.81 | -0.586 |
| GsC3H3 | 730 | 79 921.63 | 5.79 | 60.57 | 69.51 | -0.460 |
| GsC3H4 | 371 | 40 788.29 | 8.00 | 49.81 | 62.53 | -0.705 |
| GsC3H5 | 480 | 52 287.69 | 5.29 | 59.91 | 51.79 | -0.785 |
| GsC3H6 | 493 | 54 600.68 | 4.87 | 60.30 | 49.80 | -0.936 |
| GsC3H7 | 433 | 46 906.96 | 7.49 | 24.93 | 75.59 | -0.212 |
| GsC3H8 | 746 | 81 503.49 | 5.64 | 51.67 | 73.35 | -0.422 |
| GsC3H9 | 658 | 72 045.97 | 8.36 | 63.21 | 68.50 | -0.440 |
| GsC3H10 | 724 | 77 943.18 | 5.58 | 59.10 | 71.93 | -0.346 |
| GsC3H11 | 724 | 81 391.28 | 8.48 | 47.52 | 57.36 | -0.991 |
| GsC3H12 | 703 | 76 020.01 | 6.18 | 55.96 | 66.93 | -0.437 |
| GsC3H13 | 442 | 48 626.94 | 8.41 | 60.92 | 57.87 | -0.514 |
| GsC3H14 | 426 | 46 874.77 | 8.52 | 64.47 | 52.72 | -0.587 |
| GsC3H15 | 358 | 39 956.98 | 6.83 | 69.76 | 51.70 | -0.649 |
| GsC3H16 | 362 | 38 873.85 | 8.43 | 61.71 | 55.47 | -0.520 |
| GsC3H17 | 677 | 74 159.42 | 6.28 | 59.02 | 67.19 | -0.501 |
| GsC3H18 | 295 | 31 251.62 | 9.39 | 40.62 | 57.32 | -0.466 |
| GsC3H19 | 296 | 31 324.95 | 9.43 | 33.71 | 63.38 | -0.409 |
| GsC3H20 | 469 | 50 124.30 | 8.92 | 55.90 | 61.11 | -0.418 |
| GsC3H21 | 350 | 388 98.46 | 7.26 | 50.05 | 60.00 | -0.722 |
| GsC3H22 | 487 | 508 96.03 | 8.63 | 66.72 | 53.72 | -0.384 |
| GsC3H23 | 323 | 35 624.70 | 8.42 | 56.44 | 46.87 | -0.724 |
| GsC3H24 | 366 | 40 294.76 | 8.06 | 47.42 | 53.03 | -0.700 |
| GsC3H25 | 522 | 59 915.16 | 6.69 | 62.08 | 63.14 | -0.912 |
| GsC3H26 | 255 | 28 686.03 | 8.25 | 59.92 | 49.33 | -0.696 |
| GsC3H27 | 369 | 41 084.06 | 6.94 | 77.35 | 46.75 | -0.696 |
Table 2 Basic information of the C3H gene family in G. styracifolia
基因名 Gene name | 氨基酸数量 Amino acid count/aa | 相对分子质量 Relative molecular mass/Da | 等电点 Isoelectric point | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 亲水性 Hydrophilicity |
|---|---|---|---|---|---|---|
| GsC3H1 | 495 | 55 557.14 | 5.54 | 49.44 | 70.95 | -0.761 |
| GsC3H2 | 424 | 46 321.03 | 8.10 | 58.80 | 54.81 | -0.586 |
| GsC3H3 | 730 | 79 921.63 | 5.79 | 60.57 | 69.51 | -0.460 |
| GsC3H4 | 371 | 40 788.29 | 8.00 | 49.81 | 62.53 | -0.705 |
| GsC3H5 | 480 | 52 287.69 | 5.29 | 59.91 | 51.79 | -0.785 |
| GsC3H6 | 493 | 54 600.68 | 4.87 | 60.30 | 49.80 | -0.936 |
| GsC3H7 | 433 | 46 906.96 | 7.49 | 24.93 | 75.59 | -0.212 |
| GsC3H8 | 746 | 81 503.49 | 5.64 | 51.67 | 73.35 | -0.422 |
| GsC3H9 | 658 | 72 045.97 | 8.36 | 63.21 | 68.50 | -0.440 |
| GsC3H10 | 724 | 77 943.18 | 5.58 | 59.10 | 71.93 | -0.346 |
| GsC3H11 | 724 | 81 391.28 | 8.48 | 47.52 | 57.36 | -0.991 |
| GsC3H12 | 703 | 76 020.01 | 6.18 | 55.96 | 66.93 | -0.437 |
| GsC3H13 | 442 | 48 626.94 | 8.41 | 60.92 | 57.87 | -0.514 |
| GsC3H14 | 426 | 46 874.77 | 8.52 | 64.47 | 52.72 | -0.587 |
| GsC3H15 | 358 | 39 956.98 | 6.83 | 69.76 | 51.70 | -0.649 |
| GsC3H16 | 362 | 38 873.85 | 8.43 | 61.71 | 55.47 | -0.520 |
| GsC3H17 | 677 | 74 159.42 | 6.28 | 59.02 | 67.19 | -0.501 |
| GsC3H18 | 295 | 31 251.62 | 9.39 | 40.62 | 57.32 | -0.466 |
| GsC3H19 | 296 | 31 324.95 | 9.43 | 33.71 | 63.38 | -0.409 |
| GsC3H20 | 469 | 50 124.30 | 8.92 | 55.90 | 61.11 | -0.418 |
| GsC3H21 | 350 | 388 98.46 | 7.26 | 50.05 | 60.00 | -0.722 |
| GsC3H22 | 487 | 508 96.03 | 8.63 | 66.72 | 53.72 | -0.384 |
| GsC3H23 | 323 | 35 624.70 | 8.42 | 56.44 | 46.87 | -0.724 |
| GsC3H24 | 366 | 40 294.76 | 8.06 | 47.42 | 53.03 | -0.700 |
| GsC3H25 | 522 | 59 915.16 | 6.69 | 62.08 | 63.14 | -0.912 |
| GsC3H26 | 255 | 28 686.03 | 8.25 | 59.92 | 49.33 | -0.696 |
| GsC3H27 | 369 | 41 084.06 | 6.94 | 77.35 | 46.75 | -0.696 |
| 基因名 Gene name | α-螺旋 Alpha-helix/% | 延伸链 Extended strand/% | 无规则卷曲 Random coil/% | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|
| GsC3H1 | 50.71 | 6.26 | 43.03 | 细胞核 |
| GsC3H2 | 0.00 | 5.19 | 94.81 | 细胞核 |
| GsC3H3 | 26.85 | 3.70 | 69.45 | 细胞核 |
| GsC3H4 | 25.34 | 1.89 | 72.78 | 细胞核 |
| GsC3H5 | 3.12 | 4.17 | 92.71 | 细胞核 |
| GsC3H6 | 8.52 | 4.06 | 87.42 | 细胞核 |
| GsC3H7 | 2.31 | 40.18 | 57.51 | 叶绿体 |
| GsC3H8 | 33.91 | 2.41 | 63.67 | 细胞核 |
| GsC3H9 | 28.88 | 4.71 | 66.41 | 细胞核 |
| GsC3H10 | 24.59 | 3.59 | 71.82 | 细胞核 |
| GsC3H11 | 9.67 | 1.10 | 89.23 | 细胞核 |
| GsC3H12 | 17.21 | 6.12 | 76.67 | 细胞核 |
| GsC3H13 | 0.00 | 4.75 | 95.25 | 细胞核 |
| GsC3H14 | 1.64 | 4.23 | 94.13 | 细胞核 |
| GsC3H15 | 8.10 | 5.87 | 86.03 | 细胞核 |
| GsC3H16 | 0.00 | 6.35 | 93.65 | 细胞核 |
| GsC3H17 | 24.67 | 4.28 | 71.05 | 细胞核 |
| GsC3H18 | 10.17 | 5.42 | 84.41 | 过氧化物酶体 |
| GsC3H19 | 6.76 | 6.42 | 86.82 | 细胞质 |
| GsC3H20 | 0.00 | 6.18 | 93.82 | 细胞核 |
| GsC3H21 | 7.43 | 2.29 | 90.29 | 线粒体 |
| GsC3H22 | 0.00 | 3.90 | 96.10 | 细胞核 |
| GsC3H23 | 7.43 | 3.10 | 89.47 | 细胞核 |
| GsC3H24 | 17.49 | 3.01 | 79.51 | 细胞核 |
| GsC3H25 | 10.15 | 4.02 | 85.82 | 细胞核 |
| GsC3H26 | 9.41 | 8.24 | 82.35 | 细胞核 |
| GsC3H27 | 13.28 | 6.78 | 79.95 | 细胞核 |
Table 3 Secondary structure and subcellular localization analysis of the C3H gene family in G. styracifolia
| 基因名 Gene name | α-螺旋 Alpha-helix/% | 延伸链 Extended strand/% | 无规则卷曲 Random coil/% | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|
| GsC3H1 | 50.71 | 6.26 | 43.03 | 细胞核 |
| GsC3H2 | 0.00 | 5.19 | 94.81 | 细胞核 |
| GsC3H3 | 26.85 | 3.70 | 69.45 | 细胞核 |
| GsC3H4 | 25.34 | 1.89 | 72.78 | 细胞核 |
| GsC3H5 | 3.12 | 4.17 | 92.71 | 细胞核 |
| GsC3H6 | 8.52 | 4.06 | 87.42 | 细胞核 |
| GsC3H7 | 2.31 | 40.18 | 57.51 | 叶绿体 |
| GsC3H8 | 33.91 | 2.41 | 63.67 | 细胞核 |
| GsC3H9 | 28.88 | 4.71 | 66.41 | 细胞核 |
| GsC3H10 | 24.59 | 3.59 | 71.82 | 细胞核 |
| GsC3H11 | 9.67 | 1.10 | 89.23 | 细胞核 |
| GsC3H12 | 17.21 | 6.12 | 76.67 | 细胞核 |
| GsC3H13 | 0.00 | 4.75 | 95.25 | 细胞核 |
| GsC3H14 | 1.64 | 4.23 | 94.13 | 细胞核 |
| GsC3H15 | 8.10 | 5.87 | 86.03 | 细胞核 |
| GsC3H16 | 0.00 | 6.35 | 93.65 | 细胞核 |
| GsC3H17 | 24.67 | 4.28 | 71.05 | 细胞核 |
| GsC3H18 | 10.17 | 5.42 | 84.41 | 过氧化物酶体 |
| GsC3H19 | 6.76 | 6.42 | 86.82 | 细胞质 |
| GsC3H20 | 0.00 | 6.18 | 93.82 | 细胞核 |
| GsC3H21 | 7.43 | 2.29 | 90.29 | 线粒体 |
| GsC3H22 | 0.00 | 3.90 | 96.10 | 细胞核 |
| GsC3H23 | 7.43 | 3.10 | 89.47 | 细胞核 |
| GsC3H24 | 17.49 | 3.01 | 79.51 | 细胞核 |
| GsC3H25 | 10.15 | 4.02 | 85.82 | 细胞核 |
| GsC3H26 | 9.41 | 8.24 | 82.35 | 细胞核 |
| GsC3H27 | 13.28 | 6.78 | 79.95 | 细胞核 |
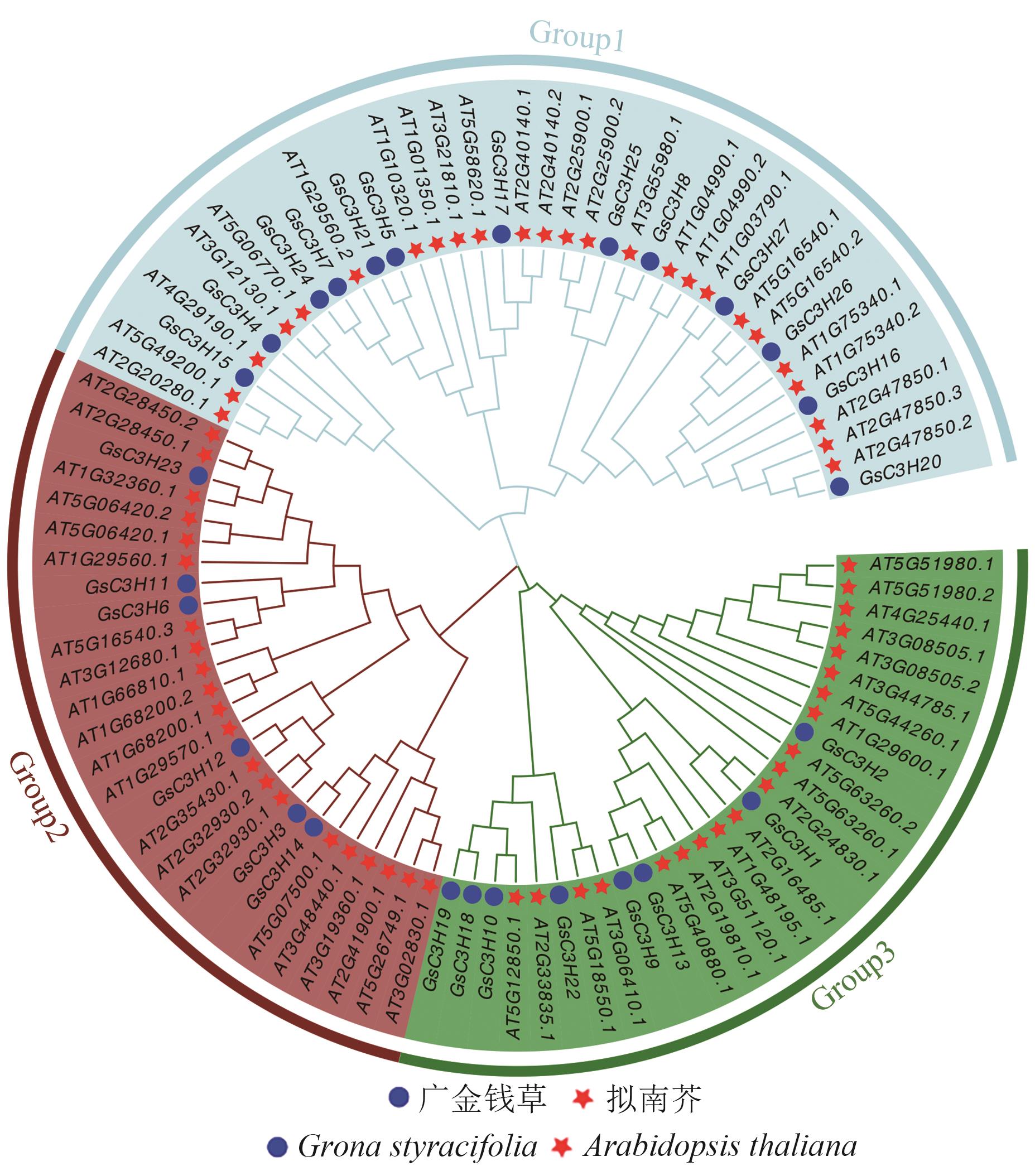
Fig. 3 Phylogenetic tree of the C3H gene family in G. styracifolia and Arabidopsis thalianaBlue, green, and brown indicate three subgroups respectively
| 基序 Motif | E值 E-value | 基序数 Sites | 序列宽度 Width/aa | 基序序列 Motif sequence |
|---|---|---|---|---|
| motif1 | 2.20E-262 | 8 | 50 | FRMFEFKVRPCSRAYSHDWTECPFVHPGENARRRDPRKYHYSCVPCPEFR |
| motif2 | 5.50E-253 | 8 | 50 | CKKGDMCEYAHGVFECWLHPAQYRTRLCKDGTSCTRRVCFFAHTPEELRP |
| motif3 | 1.90E-159 | 8 | 37 | LNYYGYPLRPGEKECPYYMRTGQCKFGATCKFHHPQP |
| motif4 | 8.70E-113 | 19 | 15 | TGTCKFGDRCKFHHP |
| motif5 | 2.00E-137 | 10 | 38 | QKEHEFPERPGQPECQYYMKTGDCKFGDKCKFHHPRWR |
| motif6 | 1.00E-104 | 7 | 39 | CILSPIGLPLRPGQPICTHYSRYGICKFGPACKFDHPMG |
| motif7 | 1.90E-84 | 9 | 35 | ESEEYPERPGEPDCTYYIRTGGCKYGKTCRFNHPP |
| motif8 | 4.60E-72 | 5 | 50 | IDEVGLWYGRRLGSKQMVYEQRTPLMVAAIYGSIDVLKYILSYSEVDVNR |
| motif9 | 1.30E-70 | 5 | 49 | CGLDKATALHCAVAGGAENAVDVVKLLLEAGADVNCVDANGNRPVDIIV |
| motif10 | 7.50E-39 | 13 | 15 | YPVRIGQPDCQYYMY |
| motif11 | 3.90E-23 | 5 | 33 | MLPGWQVQGAYGPLILPPGMVPFPGWGPYQAPM |
| motif12 | 1.30E-23 | 3 | 49 | PMSPSTNGMSHSSGCWPQPNVPALHLPGSNLQSIRLRSSLNARDIPIDD |
| motif13 | 4.00E-18 | 3 | 29 | LWDQGCEEEPVMERVESGRDIRARMFEKL |
| motif14 | 7.40E-17 | 2 | 50 | IIGKGGVNSKQICRQTGAKLSIREHESDPNLKNIELEGTFEQIKEASNMV |
| motif15 | 3.00E-16 | 5 | 26 | WGSPNGKLDWAVNGDELGKLRRSSSF |
| motif16 | 4.20E-16 | 3 | 47 | KNVEHPLLQASFGVQNSGRMSPRNMEPISPMSSRISMLAQREKQQFR |
| motif17 | 3.00E-15 | 5 | 29 | MEHLTVDTEDSFAALLELAANNDFEGFKR |
| motif18 | 6.10E-15 | 3 | 28 | VFSPTHKSAILNQFQQQQSMLSPVNTSF |
| motif19 | 6.80E-13 | 5 | 15 | LGAWLEQMYLDQLVM |
| motif20 | 2.40E-09 | 5 | 10 | FFKTKLCEKF |
Table 4 Motif information G. styracifoliaC3H gene family
| 基序 Motif | E值 E-value | 基序数 Sites | 序列宽度 Width/aa | 基序序列 Motif sequence |
|---|---|---|---|---|
| motif1 | 2.20E-262 | 8 | 50 | FRMFEFKVRPCSRAYSHDWTECPFVHPGENARRRDPRKYHYSCVPCPEFR |
| motif2 | 5.50E-253 | 8 | 50 | CKKGDMCEYAHGVFECWLHPAQYRTRLCKDGTSCTRRVCFFAHTPEELRP |
| motif3 | 1.90E-159 | 8 | 37 | LNYYGYPLRPGEKECPYYMRTGQCKFGATCKFHHPQP |
| motif4 | 8.70E-113 | 19 | 15 | TGTCKFGDRCKFHHP |
| motif5 | 2.00E-137 | 10 | 38 | QKEHEFPERPGQPECQYYMKTGDCKFGDKCKFHHPRWR |
| motif6 | 1.00E-104 | 7 | 39 | CILSPIGLPLRPGQPICTHYSRYGICKFGPACKFDHPMG |
| motif7 | 1.90E-84 | 9 | 35 | ESEEYPERPGEPDCTYYIRTGGCKYGKTCRFNHPP |
| motif8 | 4.60E-72 | 5 | 50 | IDEVGLWYGRRLGSKQMVYEQRTPLMVAAIYGSIDVLKYILSYSEVDVNR |
| motif9 | 1.30E-70 | 5 | 49 | CGLDKATALHCAVAGGAENAVDVVKLLLEAGADVNCVDANGNRPVDIIV |
| motif10 | 7.50E-39 | 13 | 15 | YPVRIGQPDCQYYMY |
| motif11 | 3.90E-23 | 5 | 33 | MLPGWQVQGAYGPLILPPGMVPFPGWGPYQAPM |
| motif12 | 1.30E-23 | 3 | 49 | PMSPSTNGMSHSSGCWPQPNVPALHLPGSNLQSIRLRSSLNARDIPIDD |
| motif13 | 4.00E-18 | 3 | 29 | LWDQGCEEEPVMERVESGRDIRARMFEKL |
| motif14 | 7.40E-17 | 2 | 50 | IIGKGGVNSKQICRQTGAKLSIREHESDPNLKNIELEGTFEQIKEASNMV |
| motif15 | 3.00E-16 | 5 | 26 | WGSPNGKLDWAVNGDELGKLRRSSSF |
| motif16 | 4.20E-16 | 3 | 47 | KNVEHPLLQASFGVQNSGRMSPRNMEPISPMSSRISMLAQREKQQFR |
| motif17 | 3.00E-15 | 5 | 29 | MEHLTVDTEDSFAALLELAANNDFEGFKR |
| motif18 | 6.10E-15 | 3 | 28 | VFSPTHKSAILNQFQQQQSMLSPVNTSF |
| motif19 | 6.80E-13 | 5 | 15 | LGAWLEQMYLDQLVM |
| motif20 | 2.40E-09 | 5 | 10 | FFKTKLCEKF |

Fig. 6 Expression analysis of C3H gene family members in different varieties and tissues of G. styracifoliaIn the figure, different lowercase letters indicate statistically significant differences (P<0.05), while the same letters indicate no significant difference
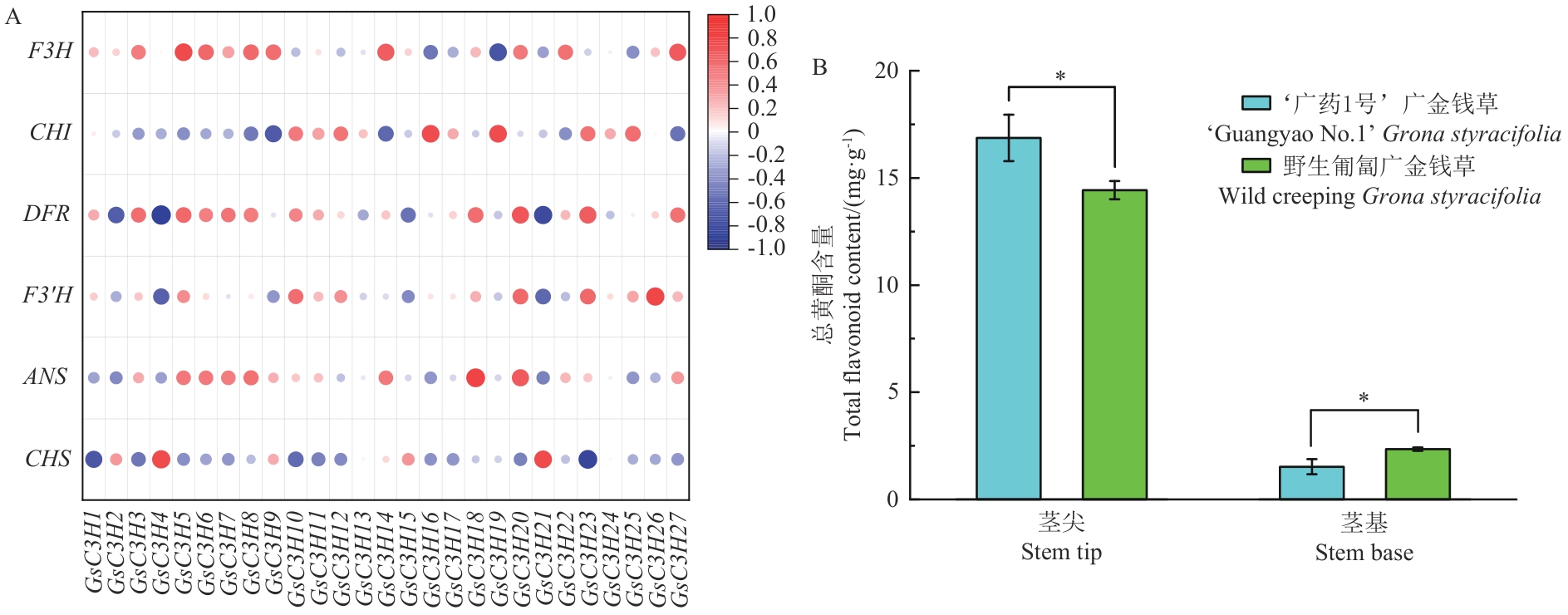
Fig. 7 Correlation analysis between G. styracifoliaC3H gene family and flavonoid biosynthesis-related genes (A) and determination of total flavonoids (B)* indicates a significant difference with P<0.05
| 1 | Xie CZ, Zhan T, Huang JQ, et al. Functional characterization of nine critical genes encoding rate-limiting enzymes in the flavonoid biosynthesis of the medicinal herb Grona styracifolia [J]. BMC Plant Biol, 2023, 23(1): 299. |
| 2 | 黄盼, 周改莲, 周文良, 等. 广金钱草的化学成分、药理作用及质量控制研究进展 [J]. 中华中医药学刊, 2021, 39(7): 135-139. |
| Huang P, Zhou GL, Zhou WL, et al. Advances in research on chemical composition, pharmacological action and quality control of Desmodium styracifolium [J]. Chin Arch Tradit Chin Med, 2021, 39(7): 135-139. | |
| 3 | Gao HY, Huang X, Lin PF, et al. Transcriptome-associated metabolomics reveals the molecular mechanism of flavonoid biosynthesis in Desmodium styracifolium (Osbeck.) Merr under abiotic stress [J]. Front Plant Sci, 2024, 15: 1431148. |
| 4 | Hou JB, Chen W, Lu HT, et al. Exploring the therapeutic mechanism of Desmodium styracifolium on oxalate crystal-induced kidney injuries using comprehensive approaches based on proteomics and network pharmacology [J]. Front Pharmacol, 2018, 9: 620. |
| 5 | Chen W, Si YC, Cheng J, et al. Metabolic and network pharmacological analyses of the therapeutic effect of Grona styracifolia on calcium oxalate-induced renal injury [J]. Front Pharmacol, 2021, 12: 652989. |
| 6 | Opryshko V, Prokhach A, Akimov O, et al. Desmodium styracifolium: Botanical and ethnopharmacological insights, phytochemical investigations, and prospects in pharmacology and pharmacotherapy [J]. Heliyon, 2024, 10(3): e25058. |
| 7 | 黄晓莉, 钟磊, 闵远洋, 等. 不同产地广金钱草种质资源综合评价 [J]. 分子植物育种, 2022, 20(23): 7960-7970. |
| Huang XL, Zhong L, Min YY, et al. Comprehensive evaluation of germplasm resources of Desmodium styraci-folium from different habitats [J]. Mol Plant Breed, 2022, 20(23): 7960-7970. | |
| 8 | 曾心宇, 唐谷华, 欧阳健明. 不同羧基含量广金钱草多糖对草酸钙晶体生长、聚集及细胞表面黏附的抑制作用 [J]. 无机化学学报, 2024, 40(8): 1563-1576. |
| Zeng XY, Tang GH, Ouyang JM. Inhibitory effect of Desmodium styracifolium polysaccharides with different content of carboxyl groups on the growth, aggregation and cell adhesion of calcium oxalate crystals [J]. Chin J Inorg Chem, 2024, 40(8): 1563-1576. | |
| 9 | 张会香, 汤霞利, 林军全, 等. 广金钱草不同极性萃取物体外降脂及降血糖活性的比较 [J]. 现代食品科技, 2023, 39(12): 192-198. |
| Zhang HX, Tang XL, Lin JQ, et al. Comparison of the in vitro lipid-lowering and hypoglycemic activities of Desmodium styracifolium extracts with different polarities [J]. Mod Food Sci Technol, 2023, 39(12): 192-198. | |
| 10 | 孙贤多. 广金钱草优良种质筛选及评价研究 [D] . 广州: 广东药科大学, 2019. |
| Sun XD. Research on the screening and evaluation of superior germplasm of Grona styracifolia [D]. Guangzhou: Guangdong Pharmaceutical University, 2019. | |
| 11 | 闵远洋. 广金钱草新品种选育研究 [D]. 广州: 广东药科大学, 2022. |
| Min YY. Study on breeding of new varieties of Desmodium styracifolium [D]. Guangzhou: Guangdong Pharmaceutical University, 2022. | |
| 12 | 尹泽群, 耿菁, 刘莹, 等. 谷子CCCH全基因家族鉴定及非生物胁迫下的表达分析 [J/OL]. 分子植物育种, 2023. . |
| Yin ZQ, Geng J, Liu Y, et al. Identification and expression analysis of the CCCH gene family in foxtail millet under abiotic stress [J/OL]. Mol Plant Breed, 2023. . | |
| 13 | 邢凯峰, 姚杏子, 周军, 等. 冷胁迫下普通油茶CCCH型锌指蛋白基因家族的鉴定和分析 [J]. 经济林研究, 2024, 42(1): 11-19. |
| Xing KF, Yao XZ, Zhou J, et al. Identification and analysis of CCCH type zinc finger protein gene family in Camellia oleifera under cold stress [J]. Non Wood For Res, 2024, 42(1): 11-19. | |
| 14 | 唐春闺, 邓兆龙, 刘琼, 等. 普通烟草CCCH类锌指蛋白家族的全基因组鉴定和表达分析 [J]. 河南农业科学, 2022, 51(4): 48-58. |
| Tang CG, Deng ZL, Liu Q, et al. Genome-wide identification and expression analysis of CCCH type zinc-finger gene family in Nicotiana tabacum [J]. J Henan Agric Sci, 2022, 51(4): 48-58. | |
| 15 | Tants JN, Oberstrass L, Weigand JE, et al. Structure and RNA-binding of the helically extended Roquin CCCH-type zinc finger [J]. Nucleic Acids Res, 2024, 52(16): 9838-9853. |
| 16 | Wang B, Fang RQ, Chen FM, et al. A novel CCCH-type zinc finger protein SAW1 activates OsGA20ox3 to regulate gibberellin homeostasis and anther development in rice [J]. J Integr Plant Biol, 2020, 62(10): 1594-1606. |
| 17 | Xie ZN, Yu GH, Lei SS, et al. CCCH protein-PvCCCH69 acted as a repressor for leaf senescence through suppressing ABA-signaling pathway [J]. Hortic Res, 2021, 8(1): 165. |
| 18 | Deng ZY, Yang ZJ, Liu XY, et al. Genome-wide identification and expression analysis of C3H zinc finger family in potato (Solanum tuberosum L.) [J]. Int J Mol Sci, 2023, 24(16): 12888. |
| 19 | Xu LA, Xiong XP, Liu TT, et al. Heterologous expression of two Brassica campestris CCCH zinc-finger proteins in Arabidopsis induces cytoplasmic foci and causes pollen abortion [J]. Int J Mol Sci, 2023, 24(23): 16862. |
| 20 | Bao PG, Sun JS, Qu GZ, et al. Identification and expression analysis of CCCH gene family and screening of key low temperature stress response gene CbuC3H24 and CbuC3H58 in Catalpa bungei [J]. BMC Genomics, 2024, 25(1): 779. |
| 21 | Teale WD, Pasternak T, Dal Bosco C, et al. Flavonol-mediated stabilization of PIN efflux complexes regulates polar auxin transport [J]. EMBO J, 2021, 40(1): e104416. |
| 22 | 郭欣慰, 黄丛林, 吴忠义, 等. 植物类黄酮生物合成的分子调控 [J]. 北方园艺, 2011(4): 204-207. |
| Guo XW, Huang CL, Wu ZY, et al. Molecular regulation of plant flavonoid biosynthesis pathway [J]. North Hortic, 2011(4): 204-207. | |
| 23 | 肖诗琪. 蜡梅CCCH型锌指蛋白基因CpC3H3在成花诱导中的功能研究 [D]. 重庆: 西南大学, 2022. |
| Xiao SQ. Study on the function of CCCH zinc finger protein gene CpC3H3 in flowering induction of Chimonanthus praecox [D]. Chongqing: Southwest University, 2022. | |
| 24 | Dong XM, Han BC, Chen JW, et al. Multiomics analyses reveal MsC3H29 positively regulates flavonoid biosynthesis to improve drought resistance of autotetraploid cultivated alfalfa (Medicago sativa L.) [J]. J Agric Food Chem, 2024, 72(25): 14448-14465. |
| 25 | Zhang M, Zhao YY, Nan TG, et al. Genome-wide analysis of Citrus medica ABC transporters reveals the regulation of fruit development by CmABCB19 and CmABCC10 [J]. Plant Physiol Biochem, 2024, 215: 109027. |
| 26 | Zeng SH, Wang ZQ, Shi DD, et al. The high-quality genome of Grona styracifolia uncovers the genomic mechanism of high levels of schaftoside, a promising drug candidate for treatment of COVID-19 [J]. Hortic Res, 2024, 11(5): uhae089. |
| 27 | 胡雨晴, 施敏, 闵远洋, 等. 6-BA和干旱胁迫下广金钱草内参基因的筛选及验证 [J]. 分子植物育种, 2023, 21(7): 2236-2244. |
| Hu YQ, Shi M, Min YY, et al. Selection and validation of internal reference genes of Desmodium styracifolium (osb.) Merr under 6-BA and drought stress [J]. Mol Plant Breed, 2023, 21(7): 2236-2244. | |
| 28 | 闫宗运. 拟南芥中含有CCCH锌指域和KH域的蛋白调控成花与衰老的机理 [D]. 北京: 中国农业大学, 2018. |
| Yan ZY. Mechanism of flowering and senescence regulation by proteins containing CCCH zinc finger and KH domains in Arabidopsis thaliana [D]. Beijing: China Agricultural University, 2018. | |
| 29 | 邓玉萍, 王倩, 张敏慧, 等. 向日葵ARF基因家族鉴定及其在花发育中的功能分析 [J]. 植物遗传资源学报, 2024, 25(11): 1967-1979. |
| Deng YP, Wang Q, Zhang MH, et al. Identification of the sunflower ARF gene family and prediction analysis of its function in flower development [J]. J Plant Genet Resour, 2024, 25(11): 1967-1979. | |
| 30 | Li DD, Yang JL, Pak S, et al. PuC3H35 confers drought tolerance by enhancing lignin and proanthocyanidin biosynthesis in the roots of Populus ussuriensis [J]. New Phytol, 2022, 233(1): 390-408. |
| 31 | Zhang SY, Liu JJ, Zhong GX, et al. Genome-wide identification and expression patterns of the C2H2-zinc finger gene family related to stress responses and catechins accumulation in Camellia sinensis[L.]O. kuntze [J]. Int J Mol Sci, 2021, 22(8): 4197. |
| [1] | LIU Tao, WANG Zhi-qi, WU Wen-bo, SHI Wen-ting, WANG Chao-nan, DU Chong, YANG Zhong-min. Identification and Expression Analysis of the GRAM Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(4): 145-155. |
| [2] | SUN Tian-guo, YI Lan, QIN Xu-yang, QIAO Meng-xue, GU Xin-ying, HAN Yi, SHA Wei, ZHANG Mei-juan, MA Tian-yi. Genome-wide Identification of the DABB Gene Family in Brassica rapa ssp. pekinensis and Expression Analysis under Saline and Alkali Stress [J]. Biotechnology Bulletin, 2025, 41(4): 156-165. |
| [3] | WANG Tian-tian, CHANG Xue-rui, HUANG Wan-yang, HUANG Jia-xin, MIAO Ru-yi, LIANG Yan-ping, WANG Jing. Identification and Analysis of GASA Gene Family in Pepper (Capsicum annuum L.) [J]. Biotechnology Bulletin, 2025, 41(4): 166-175. |
| [4] | WANG Chen, LIU Guo-mei, CHEN Chang, ZHANG Jin-long, YAO Lin, SUN Xuan, DU Chun-fang. Genome-wide Identification and Expression Analysis of CCDs Family in Brassia rapa L. [J]. Biotechnology Bulletin, 2025, 41(3): 161-170. |
| [5] | MA Tian-yi, XU Jia-jia, LU Wen-jing, WU Yan, SHA Wei, ZHANG Mei-juan, PENG Yi-fang. Expression Analysis and Resistance Identification of BrcGASA3 in Chinese Cabbage ‘Jinxiaotong’ Cultivar under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 127-138. |
| [6] | LIU Fang, DU Qian-qian, HE Hao, XIAO Gang, YAN Zhong-yuan, HAO Xiao-hua. Mechanism of miR172b/c-BnMSH7.A1 Module Responding to Cu 2+ Stress in Brassica napus [J]. Biotechnology Bulletin, 2025, 41(2): 139-149. |
| [7] | XU Yuan-meng, MAO Jiao, WANG Meng-yao, WANG Shu, REN Jiang-ling, LIU Yu-han, LIU Si-chen, QIAO Zhi-jun, WANG Rui-yun, CAO Xiao-ning. Cloning and Expression Characteristics Analysis of Millet Genes PmDEP1 and PmEP3 [J]. Biotechnology Bulletin, 2025, 41(2): 150-162. |
| [8] | JIA Zi-jian, WANG Bao-qiang, CHEN Li-fei, WANG Yi-zhen, WEI Xiao-hong, ZHAO Ying. Expression Patterns of CHX Gene Family in Quinoa in Response to NO under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 163-174. |
| [9] | QIAN Zheng-yi, WU Shao-fang, CAO Shu-yi, SONG Ya-xin, PAN Xin-feng, LI Zhao-wei, FAN Kai. Identification of the NAC Transcription Factors in Nymphaea colorata and Their Expression Analysis [J]. Biotechnology Bulletin, 2025, 41(2): 234-247. |
| [10] | XIANG Chun-fan, LI Le-song, WANG Juan, LIANG Yan-li, YANG Sheng-chao, LI Meng-fei, ZHAO Yan. Functional Identification and Expression Analysis of Cinnamonyl Alcohol Dehydrogenase AsCAD in Angelica sinensis [J]. Biotechnology Bulletin, 2025, 41(2): 295-308. |
| [11] | GE Shi-jie, LIU Yi-de, ZHANG Hua-dong, NING Qiang, ZHU Zhan-wang, WANG Shu-ping, LIU Yi-ke. Identification and Expression Analysis of Protein Disulfide Isomerase Gene Family in Wheat [J]. Biotechnology Bulletin, 2025, 41(2): 85-96. |
| [12] | LI Yu-xin, LI Miao, DU Xiao-fen, HAN Kang-ni, LIAN Shi-chao, WANG Jun. Identification and Expression Analysis of SiSAP Gene Family in Foxtail Millet(Setaria italica) [J]. Biotechnology Bulletin, 2025, 41(1): 143-156. |
| [13] | KONG Qing-yang, ZHANG Xiao-long, LI Na, ZHANG Chen-jie, ZHANG Xue-yun, YU Chao, ZHANG Qi-xiang, LUO Le. Identification and Expression Analysis of GRAS Transcription Factor Family in Rosa persica [J]. Biotechnology Bulletin, 2025, 41(1): 210-220. |
| [14] | WU Hui-qin, WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan. Identification and Expression Analysis of UGT Gene Family in Pepper [J]. Biotechnology Bulletin, 2024, 40(9): 198-211. |
| [15] | MAN Quan-cai, MENG Zi-nuo, LI Wei, CAI Xin-ru, SU Run-dong, FU Chang-qing, GAO Shun-juan, CUI Jiang-hui. Identification and Expression Analysis of AQP Gene Family in Potato [J]. Biotechnology Bulletin, 2024, 40(9): 51-63. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||