








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (2): 150-162.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0689
XU Yuan-meng1,2,3( ), MAO Jiao1,2,3, WANG Meng-yao1,2,3, WANG Shu1,2,3, REN Jiang-ling1,2,3, LIU Yu-han1,2,3, LIU Si-chen1,2,3, QIAO Zhi-jun1,2,3, WANG Rui-yun1(
), MAO Jiao1,2,3, WANG Meng-yao1,2,3, WANG Shu1,2,3, REN Jiang-ling1,2,3, LIU Yu-han1,2,3, LIU Si-chen1,2,3, QIAO Zhi-jun1,2,3, WANG Rui-yun1( ), CAO Xiao-ning1,2,3(
), CAO Xiao-ning1,2,3( )
)
Received:2024-07-18
Online:2025-02-26
Published:2025-02-28
Contact:
WANG Rui-yun, CAO Xiao-ning
E-mail:xuyuanmeng1229@163.com;wry925@126.com;caoxiaoning2008@163.com
XU Yuan-meng, MAO Jiao, WANG Meng-yao, WANG Shu, REN Jiang-ling, LIU Yu-han, LIU Si-chen, QIAO Zhi-jun, WANG Rui-yun, CAO Xiao-ning. Cloning and Expression Characteristics Analysis of Millet Genes PmDEP1 and PmEP3[J]. Biotechnology Bulletin, 2025, 41(2): 150-162.
| 引物 Primer | 引物序列 Primer sequence (5′-3′) | 功能 Primer function |
|---|---|---|
| actin | TCTGTTTTTCTCTATGTGCAGGA | 基因表达分析 Analysis of gene expression |
| PmDEP1-Q | TGCTGCTTCCATTGTTCTTGCTG | |
| PmEP3-Q | GAGCCTCCGCAAGTGGTATGG | |
| PmDEP1 | AGTGAATTCGAGCTCGGTACATGGGGGAGGAGGTGGCG | 基因克隆 Gene cloning |
| PmEP3 | AGTGAATTCGAGCTCGGTACATGGGGTCGGAAGAGTGGGAG |
Table 1 Primer sequences for PmDEP1 and PmEP3
| 引物 Primer | 引物序列 Primer sequence (5′-3′) | 功能 Primer function |
|---|---|---|
| actin | TCTGTTTTTCTCTATGTGCAGGA | 基因表达分析 Analysis of gene expression |
| PmDEP1-Q | TGCTGCTTCCATTGTTCTTGCTG | |
| PmEP3-Q | GAGCCTCCGCAAGTGGTATGG | |
| PmDEP1 | AGTGAATTCGAGCTCGGTACATGGGGGAGGAGGTGGCG | 基因克隆 Gene cloning |
| PmEP3 | AGTGAATTCGAGCTCGGTACATGGGGTCGGAAGAGTGGGAG |
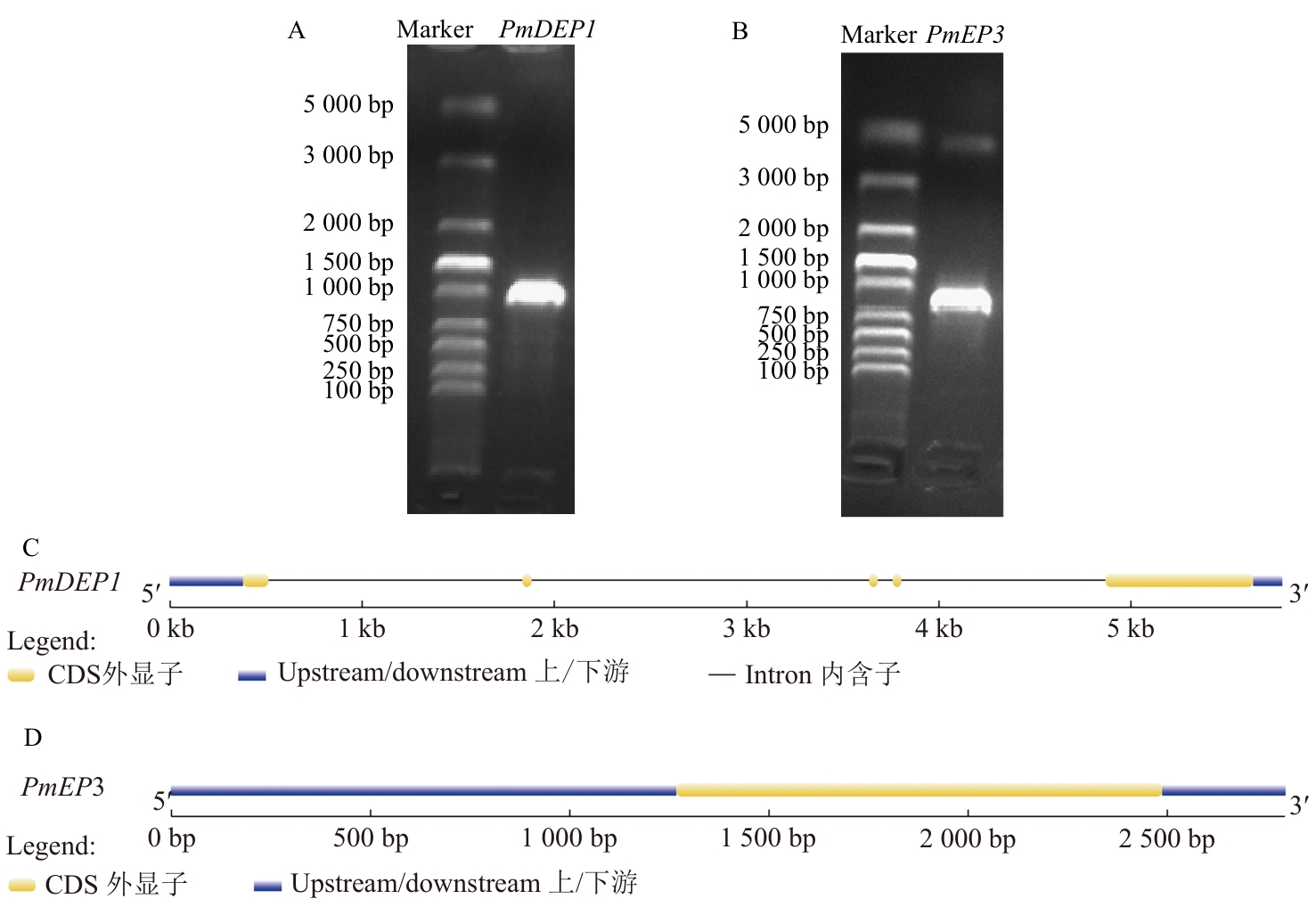
Fig. 1 Cloning and structural analysis of PmDEP1 and PmEP3 genesA: Gel electrophoresis map of PmDEP1 gene. B: Gel electrophoresis map of PmEP3 gene. C: PmDEP1 gene structure. D: PmEP3 gene structure

Fig. 2 Nucleotide and amino acid sequences and gene domains of PmDEP1 and PmEP3A: Nucleotide and amino acid sequence of PmDEP1. B: Nucleotide and amino acid sequence of PmEP3. C: PmDEP1 gene domain. D: PmEP3 gene domain
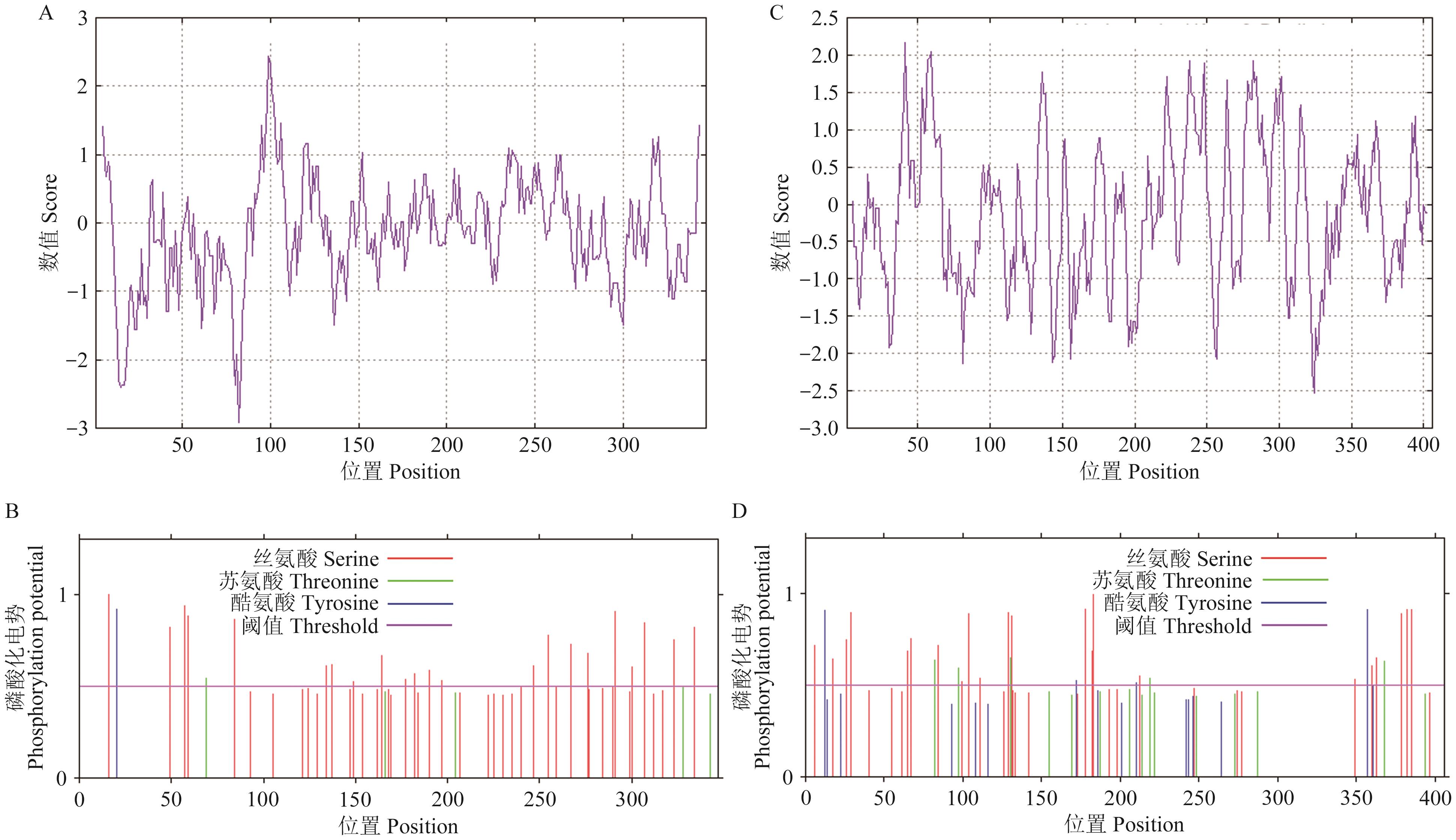
Fig. 4 Prediction of hydrophobicity/hydrophilicity and phosphorylation of PmDEP1 and PmEP3 proteinsA: Hydrophobic/hydrophilic prediction of PmDEP1 protein. B: Phosphorylation prediction of PmDEP1 protein. C: Hydrophobic/hydrophilic prediction of PmEP3 protein. D: Phosphorylation prediction of PmEP3 protein
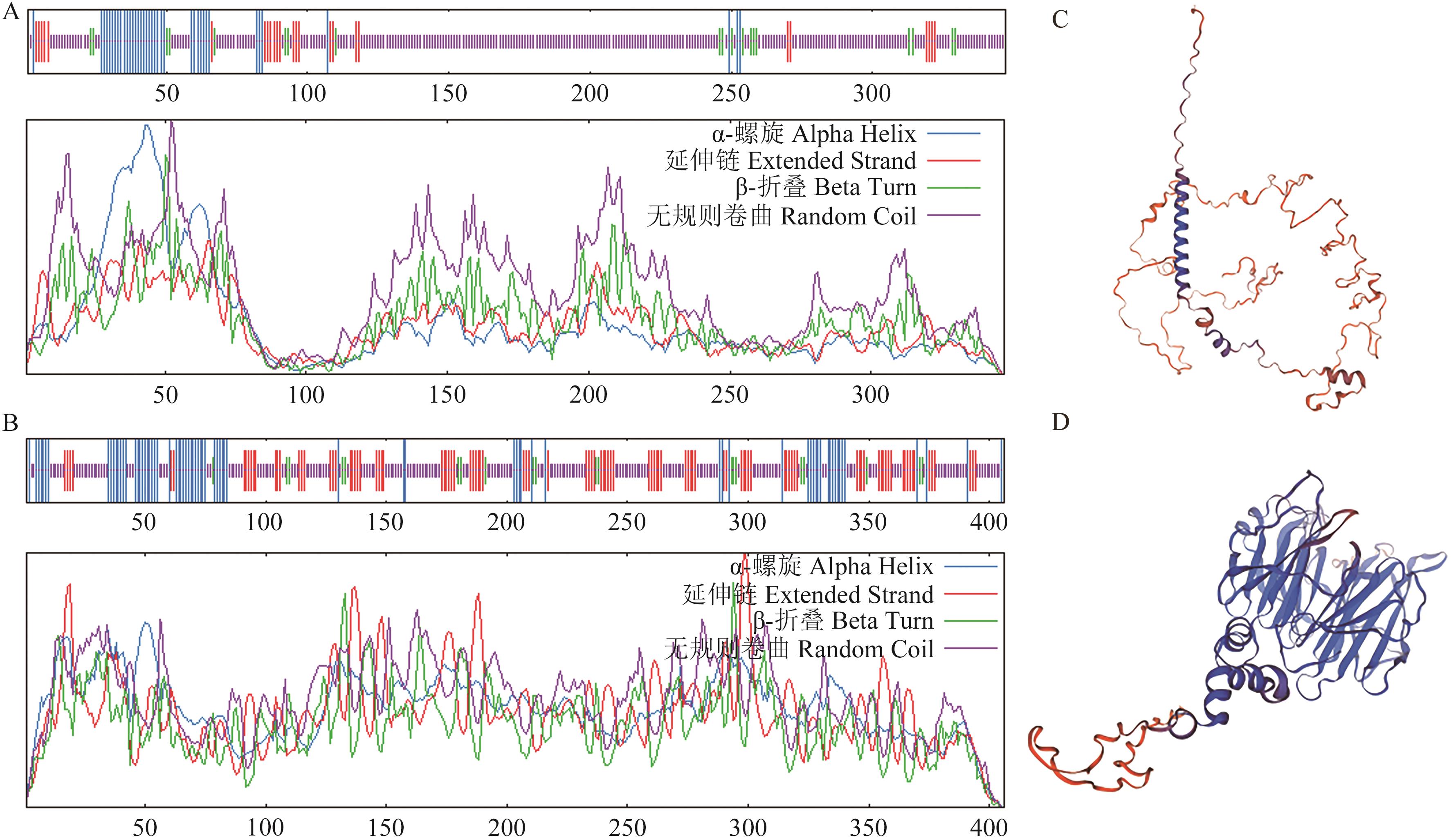
Fig. 7 Prediction of secondary and tertiary structures of PmDEP1 and PmEP3 proteinsA: Secondary structure of PmDEP1 protein. B: Secondary structure of PmEP3 protein. C: Tertiary structure of PmDEP1 protein. D: Tertiary structure of PmEP3 protein

Fig. 8 Signal peptide and transmembrane domain prediction of PmDEP1 protein and PmEP3 proteinA: Prediction of PmDEP1 protein signal peptide. B: Prediction of PmEP3 protein signal peptide. C: Prediction of PmDEP1 protein transmembrane domain. D: Prediction of PmEP3 transmembrane domain
亚细胞位置 Subcellular location | 可能性 Possibility/% | ||
|---|---|---|---|
| PmDEP1 | PmEP3 | ||
| 细胞核 | 78.3 | 26.1 | |
| 细胞质 | 8.7 | 47.8 | |
| 质膜 | 4.3 | - | |
| 过氧化物酶体 | 4.3 | - | |
| 线粒体 | 4.3 | 26.1 | |
Table 2 Prediction results of subcellular localizations for PmDEP1 and PmEP3 proteins
亚细胞位置 Subcellular location | 可能性 Possibility/% | ||
|---|---|---|---|
| PmDEP1 | PmEP3 | ||
| 细胞核 | 78.3 | 26.1 | |
| 细胞质 | 8.7 | 47.8 | |
| 质膜 | 4.3 | - | |
| 过氧化物酶体 | 4.3 | - | |
| 线粒体 | 4.3 | 26.1 | |
基因名称 Gene name | 作用元件 Functional component | 核心序列 Core sequence | 数量 Number | 功能 Function |
|---|---|---|---|---|
| PmDEP1 | CGTCA-motif | CGTCA | 1 | MeJA作用调控元件 |
| TGACG-motif | TGACG | 1 | MeJA作用调控元件 | |
| G-box | CACGAC | 4 | 光响应应答元件 | |
| chs-CMA2a | TCACTTGA | 1 | 光响应应答元件 | |
| TCA-element | CCATCTTTTT | 1 | 水杨酸作用元件 | |
| P-box | CCTTTTG | 1 | 赤霉素反应元件 | |
| ARE | AAACCA | 1 | 厌氧诱导调控元件 | |
| GC-motif | CCCCCG | 2 | 参与缺氧特异性诱导元件 | |
| MBS | CAACTG | 1 | 干旱诱导元件 | |
| CCGTCC-box | CCGTCC | 1 | 与分生组织特异性激活调节元件 | |
| STRE | AGGGG | 4 | 应激反应元件 | |
| PmEP3 | CGTCA-motif | CGTCA | 1 | MeJA作用调控元件 |
| TGACG-motif | TGACG | 1 | MeJA作用调控元件 | |
| ATCT-motif | AATCTAATCC | 1 | 光响应应答元件 | |
| TCT-motif | TCTTAC | 2 | 光响应应答元件 | |
| MRE | AACCTAA | 1 | MYB参与光反应结合位点 | |
| GARE-motif | TCTGTTG | 1 | 赤霉素反应元件 | |
| TC-rich repeats | ATTCTCTAAC | 1 | 赤霉素反应元件 | |
| AuxRR-core | GGTCCAT | 1 | 生长素反应调控元件 | |
| AAGAA-motif | GAAAGAA | 1 | 脱落酸反应性相关性元件 | |
| ARE | AAACCA | 2 | 厌氧诱导调控元件 | |
| MBS | CAACTG | 1 | 干旱诱导元件 | |
| CAT-box | GCCACT | 1 | 分生组织表达调控元件 | |
| GCN4_motif | TGAGTCA | 1 | 参与胚乳表达的顺式调控元件 |
Table 3 Analysis of cis-acting elements of PmDEP1 and PmEP3 promoters
基因名称 Gene name | 作用元件 Functional component | 核心序列 Core sequence | 数量 Number | 功能 Function |
|---|---|---|---|---|
| PmDEP1 | CGTCA-motif | CGTCA | 1 | MeJA作用调控元件 |
| TGACG-motif | TGACG | 1 | MeJA作用调控元件 | |
| G-box | CACGAC | 4 | 光响应应答元件 | |
| chs-CMA2a | TCACTTGA | 1 | 光响应应答元件 | |
| TCA-element | CCATCTTTTT | 1 | 水杨酸作用元件 | |
| P-box | CCTTTTG | 1 | 赤霉素反应元件 | |
| ARE | AAACCA | 1 | 厌氧诱导调控元件 | |
| GC-motif | CCCCCG | 2 | 参与缺氧特异性诱导元件 | |
| MBS | CAACTG | 1 | 干旱诱导元件 | |
| CCGTCC-box | CCGTCC | 1 | 与分生组织特异性激活调节元件 | |
| STRE | AGGGG | 4 | 应激反应元件 | |
| PmEP3 | CGTCA-motif | CGTCA | 1 | MeJA作用调控元件 |
| TGACG-motif | TGACG | 1 | MeJA作用调控元件 | |
| ATCT-motif | AATCTAATCC | 1 | 光响应应答元件 | |
| TCT-motif | TCTTAC | 2 | 光响应应答元件 | |
| MRE | AACCTAA | 1 | MYB参与光反应结合位点 | |
| GARE-motif | TCTGTTG | 1 | 赤霉素反应元件 | |
| TC-rich repeats | ATTCTCTAAC | 1 | 赤霉素反应元件 | |
| AuxRR-core | GGTCCAT | 1 | 生长素反应调控元件 | |
| AAGAA-motif | GAAAGAA | 1 | 脱落酸反应性相关性元件 | |
| ARE | AAACCA | 2 | 厌氧诱导调控元件 | |
| MBS | CAACTG | 1 | 干旱诱导元件 | |
| CAT-box | GCCACT | 1 | 分生组织表达调控元件 | |
| GCN4_motif | TGAGTCA | 1 | 参与胚乳表达的顺式调控元件 |
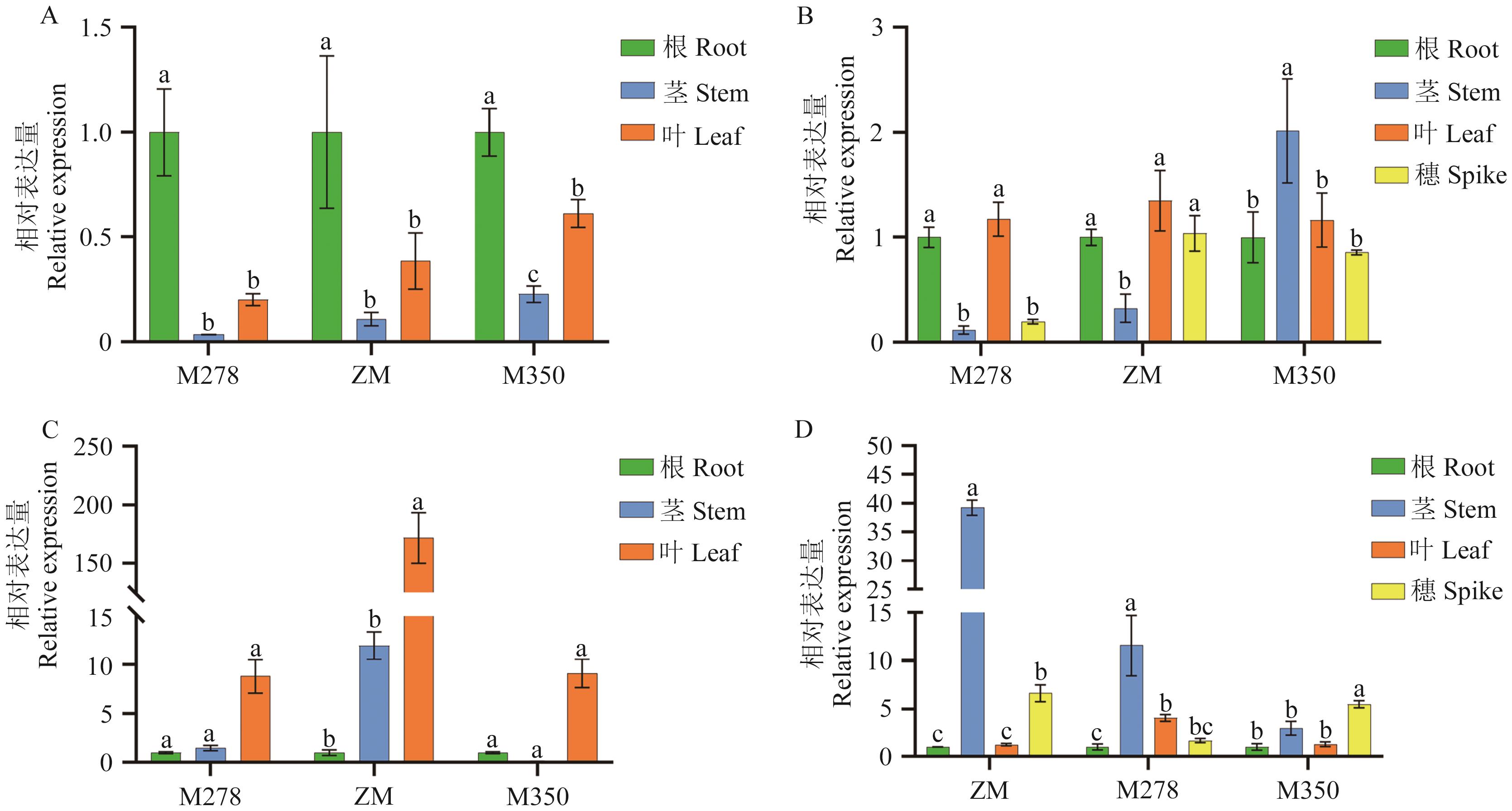
Fig. 10 Expressions of PmDEP1 and PmEP3 genes in different parts of ZM, M350 and M278 at jointing and heading stagesA: Expressions of PmDEP1 in different parts at jointing stage. B: Expressions of PmDEP1 in different parts at heading stage. C: Expressions of PmEP3 in different parts at jointing stage. D: Expressions of PmEP3 in various parts at heading stage. Different lowercase letters indicate that the expression was significantly different at the P<0.05 level
| 1 | Boukail S, Macharia M, Miculan M, et al. Genome wide association study of agronomic and seed traits in a world collection of proso millet (Panicum miliaceum L.) [J]. BMC Plant Biol, 2021, 21(1): 330. |
| 2 | Yuan YH, Liu CJ, Gao YB, et al. Proso millet (Panicum miliaceum L.): a potential crop to meet demand scenario for sustainable saline agriculture [J]. J Environ Manage, 2021, 296: 113216. |
| 3 | 王纶, 王星玉, 王海岗, 等. 山西重要黍稷种质资源品质性状的初步鉴定与评价 [J]. 植物遗传资源学报, 2017, 18(1): 61-69. |
| Wang L, Wang XY, Wang HG, et al. Preliminary appraisal of important proso mille germplasm resources quality traits in Shanxi Province [J]. J Plant Genet Resour, 2017, 18(1): 61-69. | |
| 4 | 刁现民. 禾谷类杂粮作物耐逆和栽培技术研究新进展[J]. 中国农业科学, 2019, 52(22): 3943-3949. |
| Diao XM. New research progress on stress tolerance and cultivation techniques of cereal crops[J]. Sci Agric Sin, 2019, 52(22): 3943-3949. | |
| 5 | 曹晓宁, 王君杰, 刘思辰, 等. 密穗型糜子资源的筛选评价 [J] . 现代农业科技, 2016, 18:37, 41. |
| Cao XN, Wang JJ, Liu SC, et al. Screening and evaluation of dense-spike broomcorn millet resources [J] . Mod Agric Sci Technol, 2016, 18:37, 41. | |
| 6 | 王纶, 王星玉, 乔治军, 等. 黍稷种质穗型与主要农艺性状的关系 [J]. 山西农业科学, 2013, 41(8): 789-792, 796. |
| Wang L, Wang XY, Qiao ZJ, et al. Study on broomcorn millet germplasm panicle type and its main agronomic traits [J]. J Shanxi Agric Sci, 2013, 41(8): 789-792, 796. | |
| 7 | 唐勇. 酿酒高粱SbGA2o3基因的TILLING筛选及抗旱性分析 [D]. 贵阳: 贵州大学, 2022. |
| Tang Y. TILLING screening and drought resistance analysis of SbGA2o3 gene in brewing sorghum [D]. Guiyang: Guizhou University, 2022. | |
| 8 | Fei C, Yu JH, Xu ZJ, et al. Erect panicle architecture contributes to increased rice production through the improvement of canopy structure [J]. Mol Breed, 2019, 39(9): 128. |
| 9 | Hirooka Y, Homma K, Shiraiwa T, et al. Yield and growth characteristics of erect panicle type rice (Oryza sativaL.) cultivar, Shennong265 under various crop management practices in Western Japan [J]. Plant Prod Sci, 2018, 21(1): 1-7. |
| 10 | 张喜娟, 李红娇, 李伟娟, 等. 北方直立穗型粳稻抗倒性的研究 [J]. 中国农业科学, 2009, 42(7): 2305-2313. |
| Zhang XJ, Li HJ, Li WJ, et al. The lodging resistance of erect panicle Japonica rice in northern China [J]. Sci Agric Sin, 2009, 42(7): 2305-2313. | |
| 11 | Yang YH, Shen ZY, Xu CD, et al. Genetic improvement of panicle-erectness Japonica rice toward both yield and eating and cooking quality [J]. Mol Breed, 2020, 40(5): 51. |
| 12 | Oba GC, Goneli ALD, Masetto TE, et al. Artificial drying of safflower seeds at different air temperatures: effect on the physiological potential of freshly harvested and stored seeds [J]. J Seed Sci, 2019, 41(4): 397-406. |
| 13 | Kong FN, Wang JY, Zou JC, et al. Molecular tagging and mapping of the erect panicle gene in rice [J]. Mol Breed, 2007, 19(4): 297-304. |
| 14 | Li SJ, Liu WX, Zhang XQ, et al. Roles of the Arabidopsis G protein γ subunit AGG3 and its rice homologs GS3 and DEP1 in seed and organ size control [J]. Plant Signal Behav, 2012, 7(10): 1357-1359. |
| 15 | Huang XZ, Qian Q, Liu ZB, et al. Natural variation at the DEP1 locus enhances grain yield in rice [J]. Nat Genet, 2009, 41(4): 494-497. |
| 16 | 黄勇, 胡勇, 傅向东, 等. 水稻产量性状的功能基因及其应用 [J]. 生命科学, 2016, 28(10): 1147-1155. |
| Huang Y, Hu Y, Fu XD, et al. Functional genes for grain yield related traits and their application in rice breeding [J]. Chin Bull Life Sci, 2016, 28(10): 1147-1155. | |
| 17 | Ashikari M, Sakakibara H, Lin SY, et al. Cytokinin oxidase regulates rice grain production [J]. Science, 2005, 309(5735): 741-745. |
| 18 | Wendt T, Holme I, Dockter C, et al. HvDep1 is a positive regulator of culm elongation and grain size in barley and impacts yield in an environment-dependent manner [J]. PLoS One, 2016, 11(12): e0168924. |
| 19 | Piao RH, Jiang WZ, Ham TH, et al. Map-based cloning of the ERECT PANICLE 3 gene in rice [J]. Theor Appl Genet, 2009, 119(8): 1497-1506. |
| 20 | Yu HY, Murchie EH, González-Carranza ZH, et al. Decreased photosynthesis in the erect panicle 3 (ep3) mutant of rice is associated with reduced stomatal conductance and attenuated guard cell development [J]. J Exp Bot, 2015, 66(5): 1543-1552. |
| 21 | Li M, Tang D, Wang KJ, et al. Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice [J]. Plant Biotechnol J, 2011, 9(9): 1002-1013. |
| 22 | Borna RS, Murchie EH, Pyke KA, et al. The rice EP3 and OsFBK1 E3 ligases alter plant architecture and flower development, and affect transcript accumulation of microRNA pathway genes and their targets [J]. Plant Biotechnol J, 2022, 20(2): 297-309. |
| 23 | 靳丰璐. 基于转录组测序对糜子直立穗突变体的分子机制研究 [D]. 太谷: 山西农业大学, 2022. |
| Jin FL. The study on the molecular mechanism of the erect panicle mutant in broomcorn millet based on the transcriptome sequencing analysis [D]. Taigu: Shanxi Agricultural University, 2022. | |
| 24 | Li F, Liu WB, Tang JY, et al. Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation [J]. Cell Res, 2010, 20(7): 838-849. |
| 25 | Abe Y, Mieda K, Ando T, et al. The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice [J]. Genes Genet Syst, 2010, 85(5): 327-339. |
| 26 | Qiao YL, Piao RH, Shi JX, et al. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.) [J]. Theor Appl Genet, 2011, 122(7): 1439-1449. |
| 27 | Zhu KM, Tang D, Yan CJ, et al. Erect panicle2 encodes a novel protein that regulates panicle erectness in indica rice [J]. Genetics, 2010, 184(2): 343-350. |
| 28 | Zhou Y, Zhu JY, Li ZY, et al. Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication [J]. Genetics, 2009, 183(1): 315-324. |
| 29 | 刘亚男, 夏先春, 何中虎. 普通小麦TaDep1基因克隆与特异性标记开发 [J]. 作物学报, 2013, 39(4): 589-598. |
| Liu YN, Xia XC, He ZH. Characterization of dense and erect panicle 1 gene (TaDep1) located on common wheat group 5 chromosomes and development of allele-specific markers [J]. Acta Agron Sin, 2013, 39(4): 589-598. | |
| 30 | 黄幸, 丁峰, 潘介春, 等. 龙眼DlWRKY57基因克隆与拟南芥遗传转化 [J]. 分子植物育种, 2021, 19(1): 149-157. |
| Huang X, Ding F, Pan JC, et al. Cloning and Arabidopsis genetic transformation of DlWRKY57 gene in Dimocarpus longan [J]. Mol Plant Breed, 2021, 19(1): 149-157. | |
| 31 | Liu J, Wang XY, Chen YL, et al. Identification, evolution and expression analysis of WRKY gene family in Eucommia ulmoides [J]. Genomics, 2021, 113(5): 3294-3309. |
| 32 | Wang JY, Nakazaki T, Chen SQ, et al. Identification and characterization of the erect-pose panicle gene EP conferring high grain yield in rice (Oryza sativa L.) [J]. Theor Appl Genet, 2009, 119(1): 85-91. |
| 33 | 黄先忠. 水稻直立密穗控制基因dep1的克隆和功能研究 [D]. 北京: 中国科学院研究生院, 2009. |
| Huang XZ. Cloning and functional study of the control gene dep1 for erect dense panicle in rice [D]. Beijing: Graduate University of Chinese Academy of Sciences, 2009. | |
| 34 | 吴怀通. 棉花纤维起始发育基因Li3 的图位克隆和功能验证 [D]. 南京: 南京农业大学, 2017. |
| Wu HT. Mapping cloning and functional verification of cotton fiber initiation development gene Li3 [D]. Nanjing: Nanjing Agricultural University, 2017. |
| [1] | MA Tian-yi, XU Jia-jia, LU Wen-jing, WU Yan, SHA Wei, ZHANG Mei-juan, PENG Yi-fang. Expression Analysis and Resistance Identification of BrcGASA3 in Chinese Cabbage ‘Jinxiaotong’ Cultivar under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 127-138. |
| [2] | LI Yu-xin, LI Miao, DU Xiao-fen, HAN Kang-ni, LIAN Shi-chao, WANG Jun. Identification and Expression Analysis of SiSAP Gene Family in Foxtail Millet(Setaria italica) [J]. Biotechnology Bulletin, 2025, 41(1): 143-156. |
| [3] | KONG Qing-yang, ZHANG Xiao-long, LI Na, ZHANG Chen-jie, ZHANG Xue-yun, YU Chao, ZHANG Qi-xiang, LUO Le. Identification and Expression Analysis of GRAS Transcription Factor Family in Rosa persica [J]. Biotechnology Bulletin, 2025, 41(1): 210-220. |
| [4] | SONG Bing-fang, LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi. Identification of Potato G6PDH Gene Family and Its Expression Analysis in Damaged Tubers [J]. Biotechnology Bulletin, 2024, 40(9): 104-112. |
| [5] | WU Hui-qin, WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan. Identification and Expression Analysis of UGT Gene Family in Pepper [J]. Biotechnology Bulletin, 2024, 40(9): 198-211. |
| [6] | MAN Quan-cai, MENG Zi-nuo, LI Wei, CAI Xin-ru, SU Run-dong, FU Chang-qing, GAO Shun-juan, CUI Jiang-hui. Identification and Expression Analysis of AQP Gene Family in Potato [J]. Biotechnology Bulletin, 2024, 40(9): 51-63. |
| [7] | QIAO Yan, YANG Fang, REN Pan-rong, QI Wei-liang, AN Pei-pei, LI Qian, LI Dan, XIAO Jun-fei. Cloning and Function Analysis of the ScDHNS Gene of Crotonase/Enoyl-CoA Superfamily from a Wild Potato Species [J]. Biotechnology Bulletin, 2024, 40(9): 92-103. |
| [8] | SHEN Peng, GAO Ya-Bin, DING Hong. Identification and Expression Analysis of SAT Gene Family in Potato(Solanum tuberosum L.) [J]. Biotechnology Bulletin, 2024, 40(9): 64-73. |
| [9] | LI Yi-jun, YANG Xiao-bei, XIA Lin, LUO Zhao-peng, XU Xin, YANG Jun, NING Qian-ji, WU Ming-zhu. Cloning and Functional Analysis of NtPRR37 Gene in Nicotiana tabacum L. [J]. Biotechnology Bulletin, 2024, 40(8): 221-231. |
| [10] | LI Yu-qing, WU Nan, LUO Jian-rang. Cloning and Functional Analysis of bHLH Gene Related to Anthocyanin Synthesis in Paeonia qiui [J]. Biotechnology Bulletin, 2024, 40(8): 174-185. |
| [11] | CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage [J]. Biotechnology Bulletin, 2024, 40(8): 74-82. |
| [12] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [13] | ZHOU Ran, WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma. Analysis of LncRNA Differential Expression in Mammary Tissue of Cows with Staphylococcus aureus Mastitis [J]. Biotechnology Bulletin, 2024, 40(8): 320-328. |
| [14] | ZHANG Ming-ya, PANG Sheng-qun, LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong. Identification and Expression Analysis of FAD Gene Family in Solanum lycopersicum [J]. Biotechnology Bulletin, 2024, 40(7): 150-162. |
| [15] | ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon [J]. Biotechnology Bulletin, 2024, 40(7): 163-171. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||