








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 124-138.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0390
Previous Articles Next Articles
XU Jia-yu1,2( ), ZHAO Ge3, TANG Ye2, LIU Wen-wen3, PENG Qing-zhong1(
), ZHAO Ge3, TANG Ye2, LIU Wen-wen3, PENG Qing-zhong1( ), WU Jia-he1,2(
), WU Jia-he1,2( )
)
Received:2025-04-14
Online:2025-12-26
Published:2026-01-06
Contact:
PENG Qing-zhong, WU Jia-he
E-mail:2022700533@stu.jsu.edu.cn;qzpengjsu@jsu.edu.cn;wujiahe@im.ac.cn
XU Jia-yu, ZHAO Ge, TANG Ye, LIU Wen-wen, PENG Qing-zhong, WU Jia-he. Genome-wide Identification of WRKY Gene Family in Gossypium hirsutum and Expression Analysis of WRKY44 in Fiber Development[J]. Biotechnology Bulletin, 2025, 41(12): 124-138.
| 引物名称 Primer name | 序列 Sequence (5′‒3′) |
|---|---|
| qGhhis3-F | GACACCAACCTTTGCGCGAT |
| qGhhis3-R | AGCGACTGATCCACACTTCTG |
| qGhWRKY11-F | CCGAGCTCTTGGACTCTCCTGT |
| qGhWRKY11-R | TGTTGCGCAGTTTGATTTGCGT |
| qGhWRKY14-F | AGGCAGAGCAGTCCGGAATAGG |
| qGhWRKY14-R | TGATACAAGCTTTGCCTGCGGT |
| qGhWRKY31-F | GCTCGATTACCCCACGCTTCAA |
| qGhWRKY31-R | GCCCAGTAGGCGCAGTGAATAA |
| qGhWRKY48-F | GTCTCCTCCTGTAGCAAGCCAG |
| qGhWRKY48-R | ACACAGGTGTCTTCACAACGGG |
| qGhWRKY133-F | GGTTGTCTGTGCTTCCGGCTTA |
| qGhWRKY133-R | GCTCCCTCCTTGCTTGCTTCTT |
| qGhWRKY149-F | CTTCTGCTGGGTGTCCTGTACG |
| qGhWRKY149-R | GAGCAATTGGTTGGCCATGTCG |
| qGhWRKY160-F | TGGTCAGAAGCAAGTGAAGAGTCC |
| qGhWRKY160-R | TCTCAATTACATGGCCCGTCTGA |
| qWRKY224-F | GATGGGTATCGATGGCGCAAGT |
| qWRKY224-R | TGCTTTCGGATCATGAGACGCC |
Table 1 Primer names and sequences
| 引物名称 Primer name | 序列 Sequence (5′‒3′) |
|---|---|
| qGhhis3-F | GACACCAACCTTTGCGCGAT |
| qGhhis3-R | AGCGACTGATCCACACTTCTG |
| qGhWRKY11-F | CCGAGCTCTTGGACTCTCCTGT |
| qGhWRKY11-R | TGTTGCGCAGTTTGATTTGCGT |
| qGhWRKY14-F | AGGCAGAGCAGTCCGGAATAGG |
| qGhWRKY14-R | TGATACAAGCTTTGCCTGCGGT |
| qGhWRKY31-F | GCTCGATTACCCCACGCTTCAA |
| qGhWRKY31-R | GCCCAGTAGGCGCAGTGAATAA |
| qGhWRKY48-F | GTCTCCTCCTGTAGCAAGCCAG |
| qGhWRKY48-R | ACACAGGTGTCTTCACAACGGG |
| qGhWRKY133-F | GGTTGTCTGTGCTTCCGGCTTA |
| qGhWRKY133-R | GCTCCCTCCTTGCTTGCTTCTT |
| qGhWRKY149-F | CTTCTGCTGGGTGTCCTGTACG |
| qGhWRKY149-R | GAGCAATTGGTTGGCCATGTCG |
| qGhWRKY160-F | TGGTCAGAAGCAAGTGAAGAGTCC |
| qGhWRKY160-R | TCTCAATTACATGGCCCGTCTGA |
| qWRKY224-F | GATGGGTATCGATGGCGCAAGT |
| qWRKY224-R | TGCTTTCGGATCATGAGACGCC |
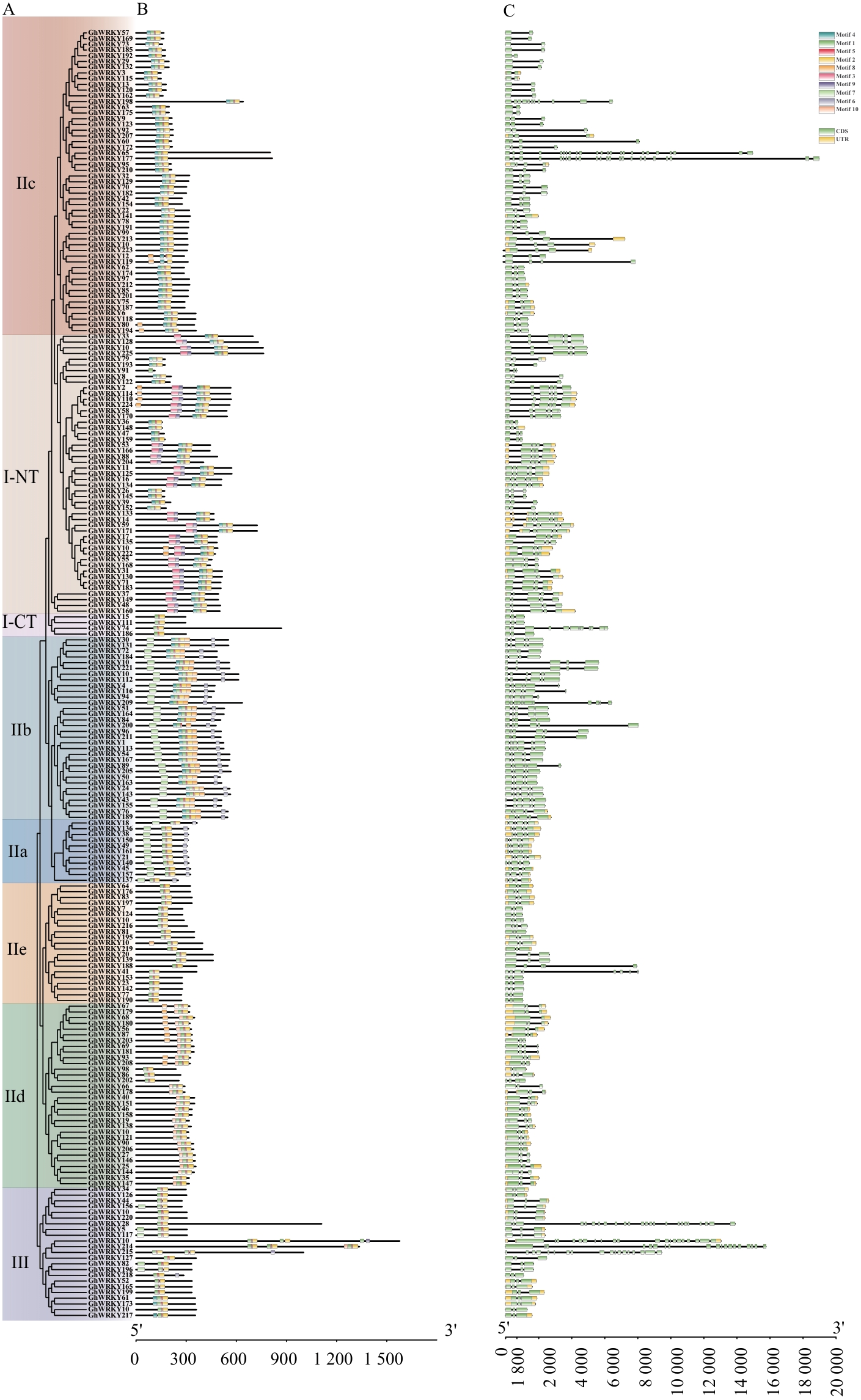
Fig. 4 Conserved motifs and gene structure analysis of GhWRKY gene family membersA: Phylogenetic tree of the GhWRKY family protein in G. hirsutum. B: Conserved motif of GhWRKY proteins in G. hirsutum. C: Gene structure of the GhWRKY gene family in G. hirsutum
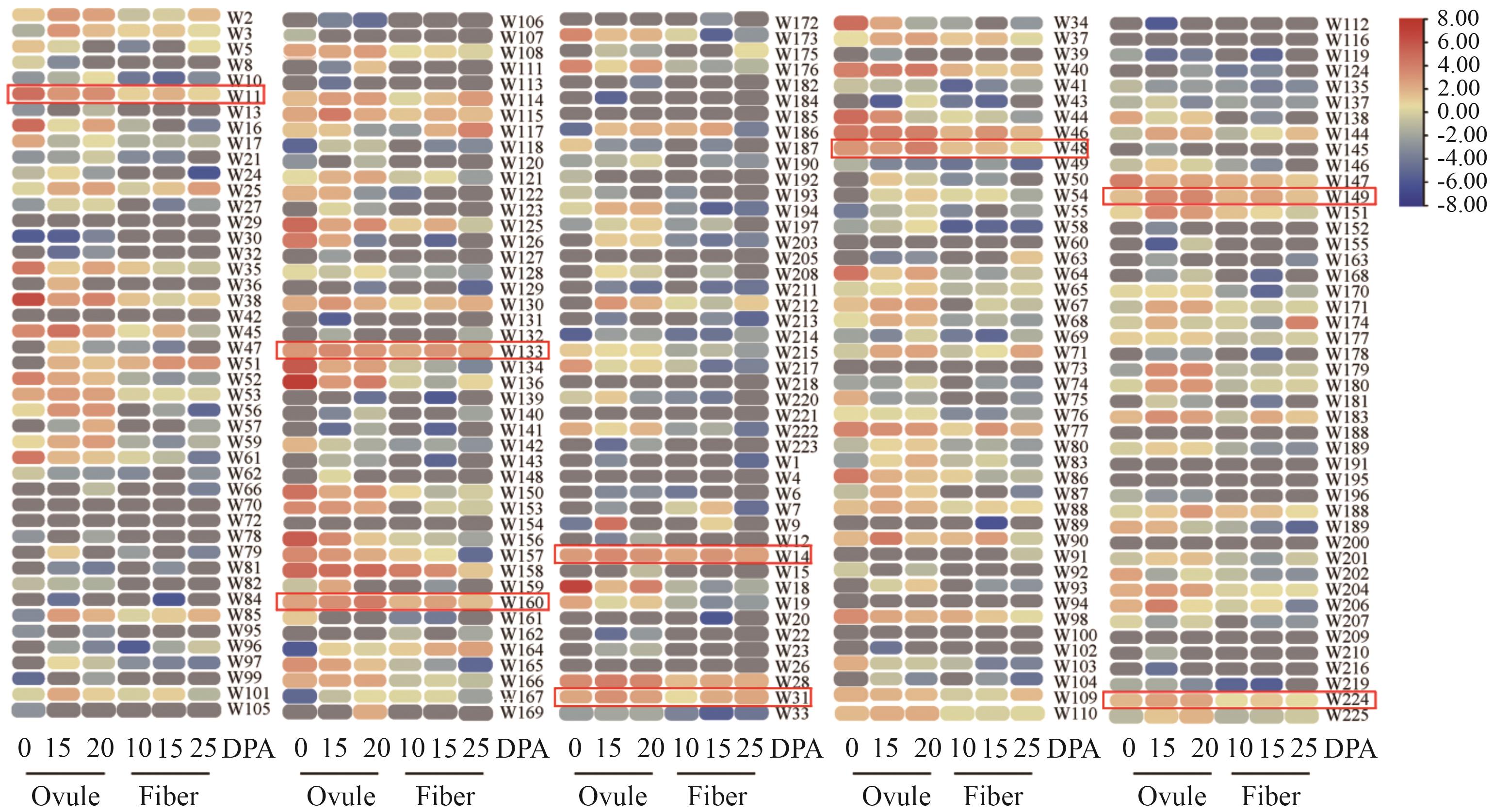
Fig. 6 Expression analysis of GhWRKYs during fiber developmentThe genes highlighted in red boxes are predominantly expressed during different developmental stages of fiber and ovule. DPA: Day post anthesis. The same below
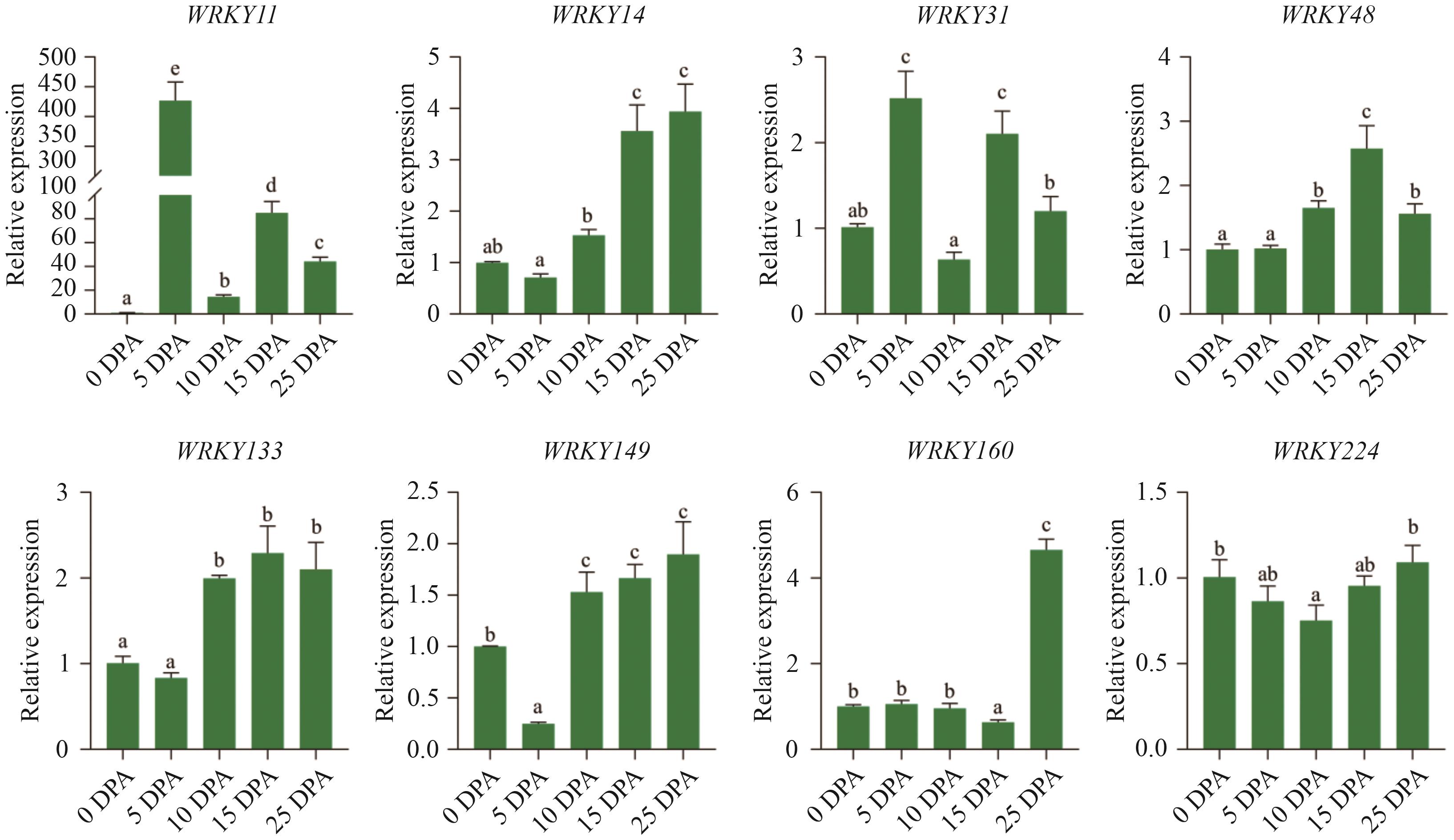
Fig. 7 Relative expressions of key GhWRKYs during different stages of cotton fiber developmentStatistical analyses were performed using the Tukey test. Different lower letters indicate significant difference of the expression of GhWRKYs at different stages of cotton fiber development (P<0.05)
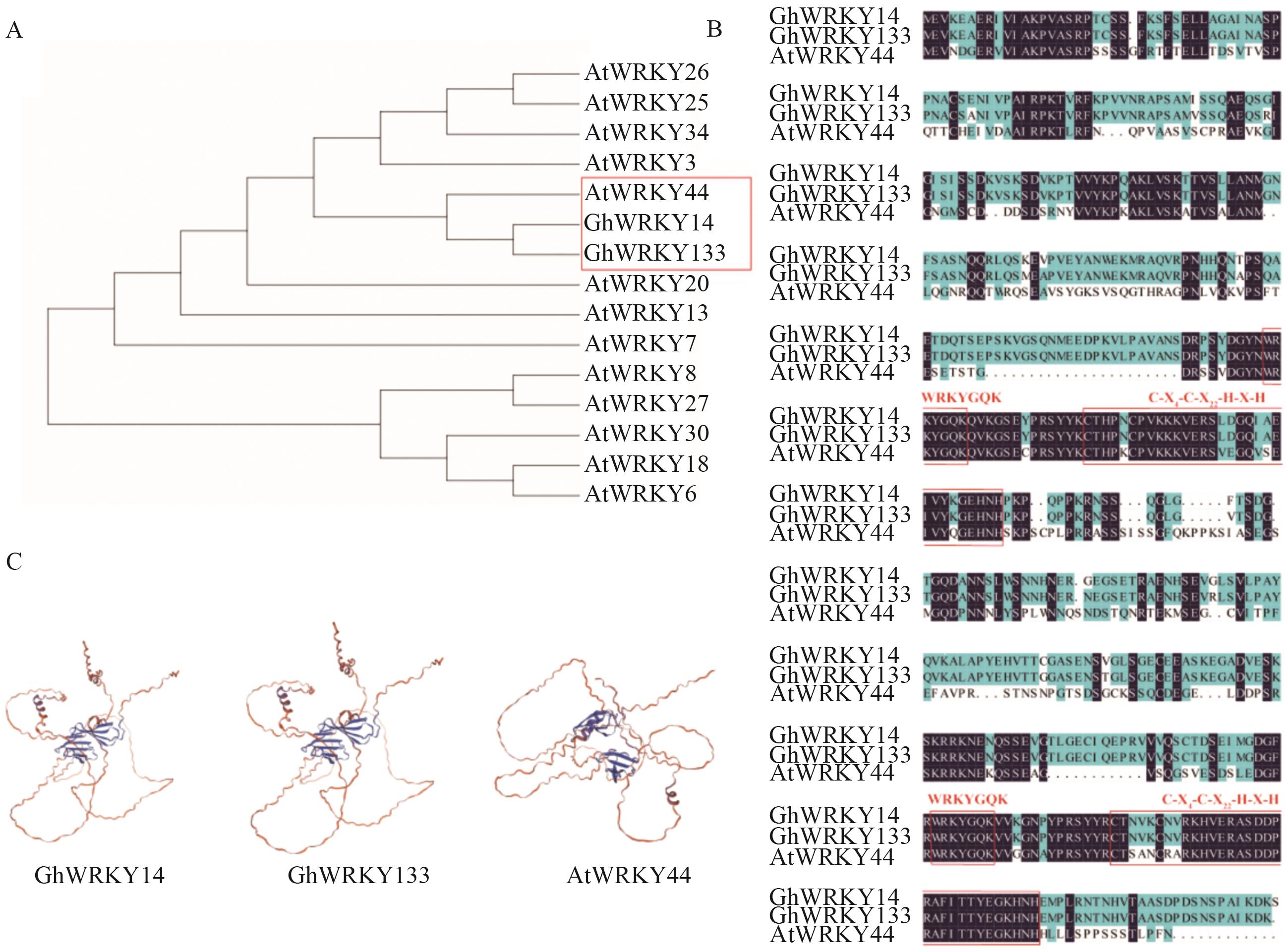
Fig. 8 Homology analysis of GhWRKY44A: Phylogenetic tree of GhWRKY14 (Gh_A04G096000.1), GhWRKY133 (Gh_D04G134100.1) and AtWRKY44 (AT2G37260). B: Amino acid sequence comparison analysis of GhWRKY14, GhWRKY133 and AtWRKY44. C: 3D structures of GhWRKY14, GhWRKY133 and AtWRKY44 proteins
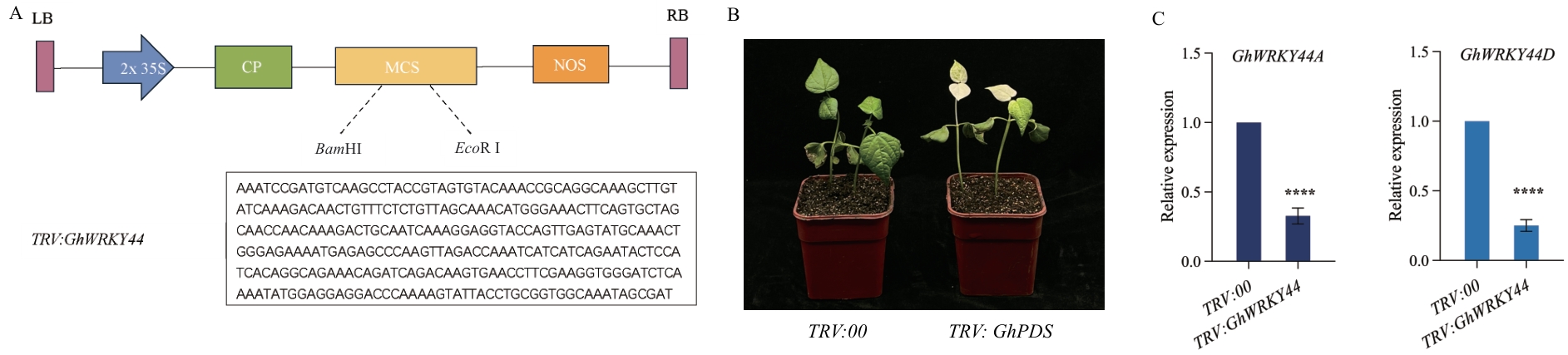
Fig. 10 Cultivating of TRV:GhWRKY44-silenced plantsA: Schematic diagram of VIGS silencing vector. B: Fourteen days after Agrobacterium infestation of cotton, TRV:GhPDS plants showed a distinct albino phenotype. C: RT-qPCR detection of GhWRKY44, silencing efficiency, n=35. GhHis3 was used as the internal reference gene, and the error line was the standard error of three biological replicates. Statistical analysis was performed using an independent variance t-test, * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001. The same below
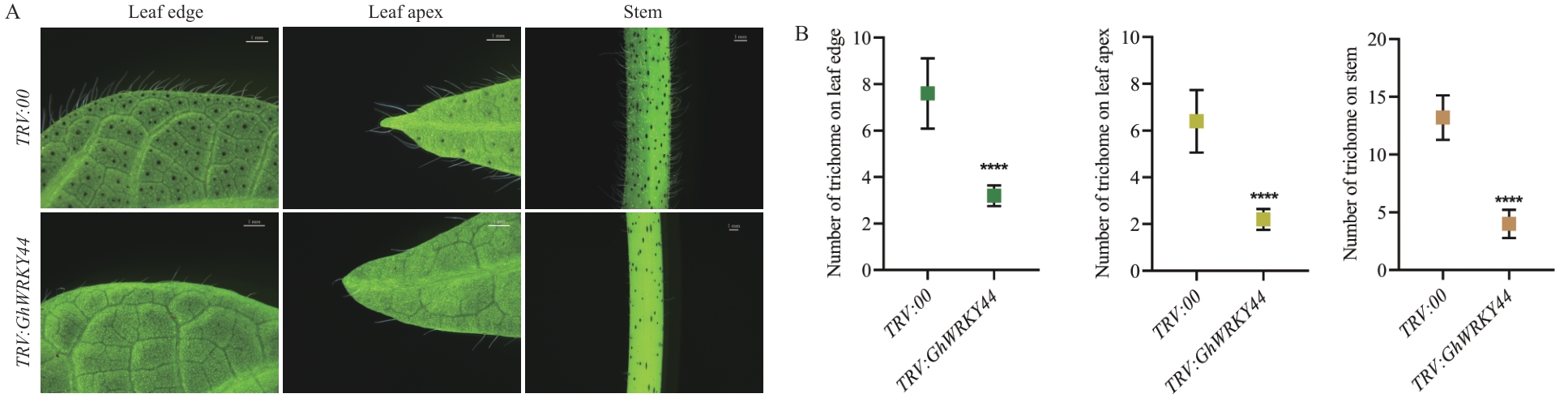
Fig. 11 Phenotypic characterization of epidermal trichomes in GhWRKY44-silenced plantsA: Trichome morphology of GhWRKY44-silenced plants. B: Statistical analysis of trichome density of GhWRKY44-silenced plants, n=15
| [1] | Qin Y, Sun ML, Li WW, et al. Single-cell RNA-seq reveals fate determination control of an individual fibre cell initiation in cotton (Gossypium hirsutum) [J]. Plant Biotechnol J, 2022, 20(12): 2372-2388. |
| [2] | Tian HD, Wang QL, Yan XY, et al. The disruptions of sphingolipid and sterol metabolism in the short fiber of Ligon-lintless-1 mutant revealed obesity impeded cotton fiber elongation and secondary cell wall deposition [J]. Int J Mol Sci, 2025, 26(3): 1375. |
| [3] | Kim HJ, Triplett BA. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis [J]. Plant Physiol, 2001, 127(4): 1361-1366. |
| [4] | Haigler CH, Betancur L, Stiff MR, et al. Cotton fiber: a powerful single-cell model for cell wall and cellulose research [J]. Front Plant Sci, 2012, 3: 104. |
| [5] | Wu N, Yang J, Wang GN, et al. Novel insights into water-deficit-responsive mRNAs and lncRNAs during fiber development in Gossypium hirsutum [J]. BMC Plant Biol, 2022, 22(1): 6. |
| [6] | Sun XW, Qin AZ, Wang XX, et al. Spatiotemporal transcriptome and metabolome landscapes of cotton fiber during initiation and early development [J]. Nat Commun, 2025, 16(1): 858. |
| [7] | Wani SH, Anand S, Singh B, et al. WRKY transcription factors and plant defense responses: latest discoveries and future prospects [J]. Plant Cell Rep, 2021, 40(7): 1071-1085. |
| [8] | Song H, Cao YP, Zhao LG, et al. Review: WRKY transcription factors: Understanding the functional divergence [J]. Plant Sci, 2023, 334: 111770. |
| [9] | Eulgem T, Rushton PJ, Robatzek S, et al. The WRKY superfamily of plant transcription factors [J]. Trends Plant Sci, 2000, 5(5): 199-206. |
| [10] | Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1 that recognizes SP8 sequences in the 5' upstream regions of genes coding for sporamin and beta-amylase from sweet potato [J]. Mol Gen Genet, 1994, 244(6): 563-571. |
| [11] | Ülker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function [J]. Curr Opin Plant Biol, 2004, 7(5): 491-498. |
| [12] | Sahebi M, Hanafi MM, Rafii MY, et al. Improvement of drought tolerance in rice (Oryza sativa L.): genetics, genomic tools, and the WRKY gene family [J]. Biomed Res Int, 2018, 2018: 3158474. |
| [13] | Hu WJ, Ren QY, Chen YL, et al. Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses [J]. BMC Plant Biol, 2021, 21(1): 427. |
| [14] | Xing LN, Zhang YF, Ge MR, et al. Identification of WRKY gene family in Dioscorea opposita Thunb. reveals that DoWRKY71 enhanced the tolerance to cold and ABA stress [J]. Peer J, 2024, 12: e17016. |
| [15] | Lei L, Dong K, Liu SW, et al. Genome-wide identification of the WRKY gene family in blueberry (Vaccinium spp.) and expression analysis under abiotic stress [J]. Front Plant Sci, 2024, 15: 1447749. |
| [16] | Xiong R, Peng Z, Zhou H, et al. Correction: Genome-wide identification, structural characterization and gene expression analysis of the WRKY transcription factor family in pea (Pisum sativum L.) [J]. BMC Plant Biol, 2024, 24(1): 141. |
| [17] | Zhao H, Wang S, Chen S, et al. Phylogenetic and stress-responsive expression analysis of 20 WRKY genes in Populus simonii × Populus nigra [J]. Gene, 2015, 565(1): 130-139. |
| [18] | Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2 a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor [J]. Plant Cell, 2002, 14(6): 1359-1375. |
| [19] | Liu R, Xu YH, Jiang SC, et al. Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function [J]. J Exp Bot, 2013, 64(18): 5443-5456. |
| [20] | Ding ZJ, Yan JY, Li GX, et al. WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA [J]. Plant J, 2014, 79(5): 810-823. |
| [21] | Lei RH, Ma ZB, Yu DQ. WRKY2/34-VQ20 modules in Arabidopsis thaliana negatively regulate expression of a trio of related MYB transcription factors during pollen development [J]. Front Plant Sci, 2018, 9: 331. |
| [22] | Li W, Wang HP, Yu DQ. Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions [J]. Mol Plant, 2016, 9(11): 1492-1503. |
| [23] | Ma ZB, Li W, Wang HP, et al. WRKY transcription factors WRKY12 and WRKY13 interact with SPL10 to modulate age-mediated flowering [J]. J Integr Plant Biol, 2020, 62(11): 1659-1673. |
| [24] | Zhang LP, Chen LG, Yu DQ. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering [J]. Plant Physiol, 2018, 176(1): 790-803. |
| [25] | Rishmawi L, Pesch M, Juengst C, et al. Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis [J]. Plant Physiol, 2014, 165(1): 186-195. |
| [26] | Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense [J]. Genes Dev, 2002, 16(9): 1139-1149. |
| [27] | Doll J, Muth M, Riester L, et al. Arabidopsis thaliana WRKY25 transcription factor mediates oxidative stress tolerance and regulates senescence in a redox-dependent manner [J]. Front Plant Sci, 2020, 10: 1734. |
| [28] | Chen LG, Xiang SY, Chen YL, et al. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence [J]. Mol Plant, 2017, 10(9): 1174-1189. |
| [29] | Jiang YJ, Yu DQ. The WRKY57 transcription factor affects the expression of jasmonate ZIM-domain genes transcriptionally to compromise Botrytis cinerea resistance [J]. Plant Physiol, 2016, 171(4): 2771-2782. |
| [30] | Wu XL, Shiroto Y, Kishitani S, et al. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter [J]. Plant Cell Rep, 2009, 28(1): 21-30. |
| [31] | He GH, Xu JY, Wang YX, et al. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis [J]. BMC Plant Biol, 2016, 16(1): 116. |
| [32] | Li FJ, Li MY, Wang P, et al. Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59 [J]. New Phytol, 2017, 215(4): 1462-1475. |
| [33] | Li SJ, Fu QT, Chen LG, et al. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance [J]. Planta, 2011, 233(6): 1237-1252. |
| [34] | Zhang Y, Yu HJ, Yang XY, et al. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner [J]. Plant Physiol Biochem, 2016, 108: 478-487. |
| [35] | Yokotani N, Sato Y, Tanabe S, et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance [J]. J Exp Bot, 2013, 64(16): 5085-5097. |
| [36] | Wang NN, Li Y, Chen YH, et al. Phosphorylation of WRKY16 by MPK3-1 is essential for its transcriptional activity during fiber initiation and elongation in cotton (Gossypium hirsutum) [J]. Plant Cell, 2021, 33(8): 2736-2752. |
| [37] | Yang L, Qin WQ, Wei X, et al. Regulatory networks of coresident subgenomes during rapid fiber cell elongation in upland cotton [J]. Plant Commun, 2024, 5(12): 101130. |
| [38] | Yang DJ, Liu YY, Cheng HL, et al. Identification of the group ⅢWRKY subfamily and the functional analysis of GhWRKY53 in Gossypium hirsutum L [J]. Plants, 2021, 10(6): 1235. |
| [39] | Wang HP, Chen WQ, Xu ZY, et al. Functions of WRKYs in plant growth and development [J]. Trends Plant Sci, 2023, 28(6): 630-645. |
| [40] | Sun PW, Xu YH, Yu CC, et al. WRKY44 represses expression of the wound-induced sesquiterpene biosynthetic gene ASS1 in Aquilaria sinensis [J]. J Exp Bot, 2020, 71(3): 1128-1138. |
| [41] | Peng YY, Thrimawithana AH, Cooney JM, et al. The proanthocyanin-related transcription factors MYBC1 and WRKY44 regulate branch points in the kiwifruit anthocyanin pathway [J]. Sci Rep, 2020, 10(1): 14161. |
| [42] | Han Y, Zhang X, Wang W, et al. Correction: the suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana [J]. PLoS One, 2015, 10(4): e0124854. |
| [43] | Li QY, Yin M, Li YP, et al. Expression of Brassica napus TTG2 a regulator of trichome development, increases plant sensitivity to salt stress by suppressing the expression of auxin biosynthesis genes [J]. J Exp Bot, 2015, 66(19): 5821-5836. |
| [44] | Wen TY, Xu X, Ren AP, et al. Genome-wide identification of terpenoid synthase family genes in Gossypium hirsutum and functional dissection of its subfamily cadinene synthase A in gossypol synthesis [J]. Front Plant Sci, 2023, 14: 1162237. |
| [45] | Bencke-Malato M, Cabreira C, Wiebke-Strohm B, et al. Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection [J]. BMC Plant Biol, 2014, 14: 236. |
| [46] | Shi GF, Liu GL, Liu HJ, et al. WRKY transcriptional factor IlWRKY70 from Iris laevigata enhances drought and salinity tolerances in Nicotiana tabacum [J]. Int J Mol Sci, 2023, 24(22): 16174. |
| [47] | Zhang JW, Huang DZ, Zhao XJ, et al. Drought-responsive WRKY transcription factor genes IgWRKY50 and IgWRKY32 from Iris germanica enhance drought resistance in transgenic Arabidopsis [J]. Front Plant Sci, 2022, 13: 983600. |
| [48] | Chen JN, Nolan TM, Ye HX, et al. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses [J]. Plant Cell, 2017, 29(6): 1425-1439. |
| [49] | Shim JS, Jung C, Lee S, et al. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling [J]. Plant J, 2013, 73(3): 483-495. |
| [50] | Turck F, Zhou AF, Somssich IE. Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene PcPR1-1 in Parsley [J]. Plant Cell, 2004, 16(10): 2573-2585. |
| [1] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [2] | GONG Hui-ling, XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping. Identification of Laccase (LAC) Gene Family in Potato (Solanum tuberosum L.) and Its Expression Analysis under Salt Stresses [J]. Biotechnology Bulletin, 2025, 41(9): 82-93. |
| [3] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [4] | BAI Yu-guo, LI Wan-di, LIANG Jian-ping, SHI Zhi-yong, LU Geng-long, LIU Hong-jun, NIU Jing-ping. Growth-promoting Mechanism of Trichoderma harzianum T9131 on Astragalus membranaceus Seedlings [J]. Biotechnology Bulletin, 2025, 41(8): 175-185. |
| [5] | HUA Wen-ping, LIU Fei, HAO Jia-xin, CHEN Chen. Identification and Expression Patterns Analysis of ADH Gene Family in Salvia miltiorrhiza [J]. Biotechnology Bulletin, 2025, 41(8): 211-219. |
| [6] | REN Rui-bin, SI Er-jing, WAN Guang-you, WANG Jun-cheng, YAO Li-rong, ZHANG Hong, MA Xiao-le, LI Bao-chun, WANG Hua-jun, MENG Ya-xiong. Identification and Expression Analysis of GH17 Gene Family of Pyrenophora graminea [J]. Biotechnology Bulletin, 2025, 41(8): 146-154. |
| [7] | ZENG Dan, HUANG Yuan, WANG Jian, ZHANG Yan, LIU Qing-xia, GU Rong-hui, SUN Qing-wen, CHEN Hong-yu. Genome-wide Identification and Expression Analysis of bZIP Transcription Factor Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2025, 41(8): 197-210. |
| [8] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [9] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [10] | HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare [J]. Biotechnology Bulletin, 2025, 41(7): 106-116. |
| [11] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [12] | QU Mei-ling, ZHOU Si-min, ZHANG Jing-yu, HE Jia-wei, ZHU Jia-yuan, LIU Xiao-rong, TONG Qiao-zhen, ZHOU Ri-bao, LIU Xiang-dan. Identification and Expression Analysis of bHLH Transcription Gene Family in Lonicera macranthoides [J]. Biotechnology Bulletin, 2025, 41(6): 256-268. |
| [13] | WANG Miao-miao, ZHAO Xiang-long, WANG Zhao-ming, LIU Zhi-peng, YAN Long-feng. Identification of TCP Gene Family in Medicago ruthenica and Their Expression Pattern Analysis under Drought Stress [J]. Biotechnology Bulletin, 2025, 41(6): 179-190. |
| [14] | LIU Xin, WANG Jia-wen, LI Jin-wei, MOU Ce, YANG Pan-pan, MING Jun, XU Lei-feng. Cloning and Expression Analysis of Three LdBBXs in Lilium davidii var. willmottiae [J]. Biotechnology Bulletin, 2025, 41(5): 186-196. |
| [15] | ZHAO Jing, GUO Qian, LI Rui-qi, LEI Ying-yang, YUE Ai-qin, ZHAO Jin-zhong, YIN Cong-cong, DU Wei-jun, NIU Jing-ping. Sequence Analysis and Induced Expression Analysis of GmGST Gene Cluster Genes in Soybean [J]. Biotechnology Bulletin, 2025, 41(5): 129-140. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||