








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (11): 272-281.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0423
CHEN Lin-lin( ), SU Zeng-qing, JI Xiao-ya, LIU Xin-yue, LIU Jia-yao, ZHANG Shi-yu, XING Xiao-ping, LI Hong-lian(
), SU Zeng-qing, JI Xiao-ya, LIU Xin-yue, LIU Jia-yao, ZHANG Shi-yu, XING Xiao-ping, LI Hong-lian( )
)
Received:2025-04-22
Online:2025-11-26
Published:2025-12-09
Contact:
LI Hong-lian
E-mail:llchensky@163.com;honglianli@sina.com
CHEN Lin-lin, SU Zeng-qing, JI Xiao-ya, LIU Xin-yue, LIU Jia-yao, ZHANG Shi-yu, XING Xiao-ping, LI Hong-lian. Metacaspase-like FpMca3 Involved in Pathogenicity of Fusarium pseudograminearum[J]. Biotechnology Bulletin, 2025, 41(11): 272-281.
| Primer | Primer sequence (5′-3′) |
|---|---|
| FpMca1-F | ATGTCGCAATACCCAGGC |
| FpMca1-R | CTAATACTTGGTCAATGGC |
| FpMca2-F | ATGTCCTACTTTCCAGGTC |
| FpMca2-R | CATAACAAACAAGAGGTCGG |
| FpMca3-F | ATGTCCAACAAGGCCGTC |
| FpMca3-R | TCACACATTCGTAGGTACC |
| FpMca4-F | ATGACAAACCAACCGACTCATC |
| FpMca4-R | CTAAAGTACCGGTATCGAGGT |
| F1 | ACAAGCGAATGCGAGTCGGTCAG |
| R1 | TTGACCTCCACTAGCTCCAGCCAAGCCGATTGGTCTGACCATGTAAT |
| F2 | ATAGAGTAGATGCCGACCGCGGGTTCAATTAATTGCAGAATTCTC |
| R2 | TACTTCCAAAGTATATCACTGC |
| UP-F | GAGTGTGAGAGGATGTTGTC |
| H855R | GCTGATCTGACCAGTTGC |
| H856F | GTCGATGCGACGCAATCGT |
| Down-R | GCTGAATATCCTTTGCTGTTG |
| H850F | TTCCTCCCTTTATTTCAGATTCAA |
| H852R | ATGTTGGCGACCTCGTATTGG |
| G-F | CTAGCAATATCACCCTCATC |
| G-R | GGAGACTTTGGCAGTGTTC |
| Com-F | CTATAGGGCGAATTGGGTACCGAATTGATCTGTATACTGTAG |
| Com-R | GCAGGCATGCAAGCTTATCGATCACATTCGTAGGTACCATTTC |
| RT-F | GAGCTGGTCAACAATGGA |
| RT-R | CTTGTAGCCCCTTGAGTTC |
| FpTEF1-RTF | TCACCACTGAAGTCAAGTCC |
| FpTEF1-RTR | ACCAGCGACGTTACCACGTC |
Table 1 Sequences of primers used in this study
| Primer | Primer sequence (5′-3′) |
|---|---|
| FpMca1-F | ATGTCGCAATACCCAGGC |
| FpMca1-R | CTAATACTTGGTCAATGGC |
| FpMca2-F | ATGTCCTACTTTCCAGGTC |
| FpMca2-R | CATAACAAACAAGAGGTCGG |
| FpMca3-F | ATGTCCAACAAGGCCGTC |
| FpMca3-R | TCACACATTCGTAGGTACC |
| FpMca4-F | ATGACAAACCAACCGACTCATC |
| FpMca4-R | CTAAAGTACCGGTATCGAGGT |
| F1 | ACAAGCGAATGCGAGTCGGTCAG |
| R1 | TTGACCTCCACTAGCTCCAGCCAAGCCGATTGGTCTGACCATGTAAT |
| F2 | ATAGAGTAGATGCCGACCGCGGGTTCAATTAATTGCAGAATTCTC |
| R2 | TACTTCCAAAGTATATCACTGC |
| UP-F | GAGTGTGAGAGGATGTTGTC |
| H855R | GCTGATCTGACCAGTTGC |
| H856F | GTCGATGCGACGCAATCGT |
| Down-R | GCTGAATATCCTTTGCTGTTG |
| H850F | TTCCTCCCTTTATTTCAGATTCAA |
| H852R | ATGTTGGCGACCTCGTATTGG |
| G-F | CTAGCAATATCACCCTCATC |
| G-R | GGAGACTTTGGCAGTGTTC |
| Com-F | CTATAGGGCGAATTGGGTACCGAATTGATCTGTATACTGTAG |
| Com-R | GCAGGCATGCAAGCTTATCGATCACATTCGTAGGTACCATTTC |
| RT-F | GAGCTGGTCAACAATGGA |
| RT-R | CTTGTAGCCCCTTGAGTTC |
| FpTEF1-RTF | TCACCACTGAAGTCAAGTCC |
| FpTEF1-RTR | ACCAGCGACGTTACCACGTC |
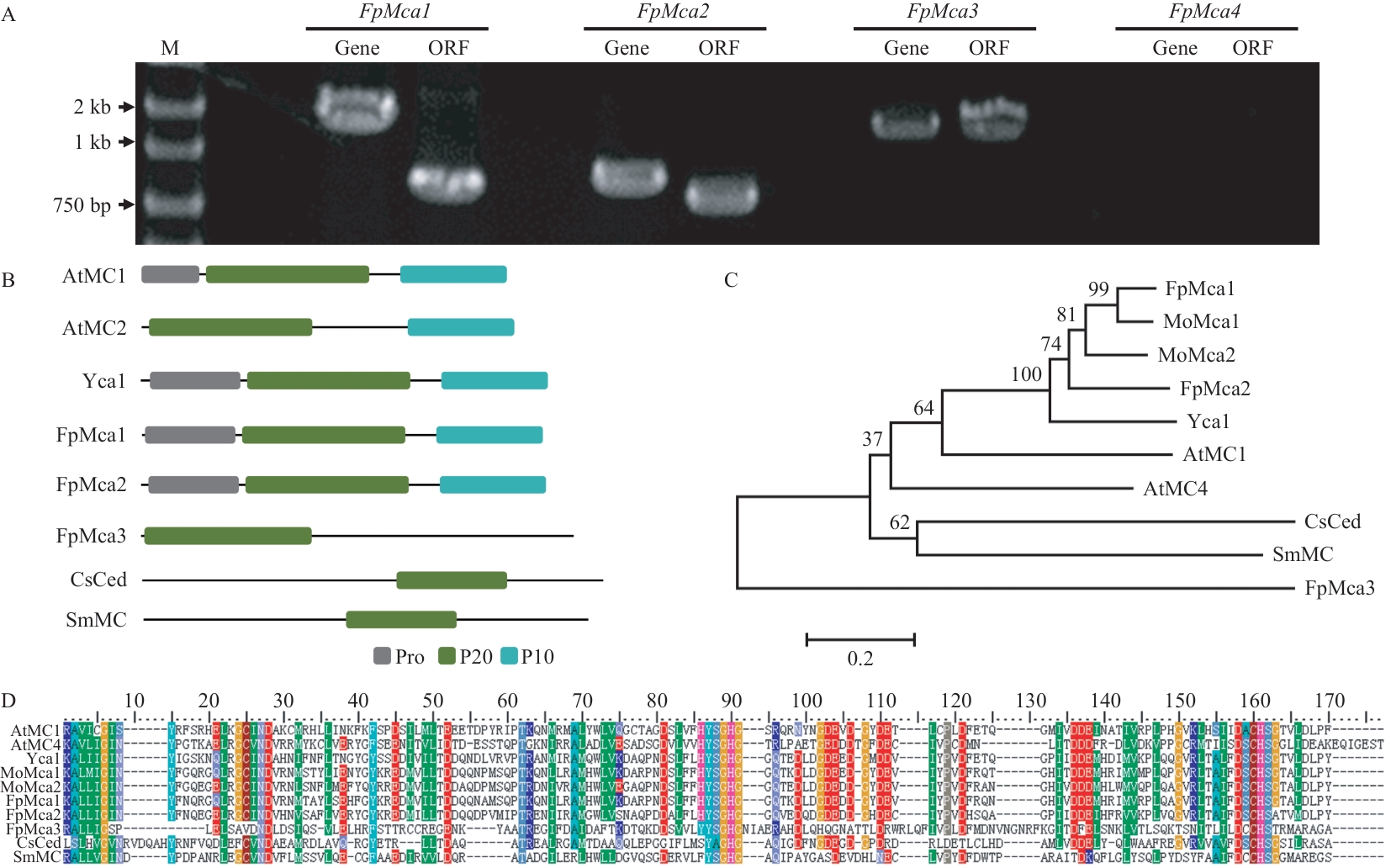
Fig. 1 Cloning and analysis of FpMcasA: Cloning of FpMca1-4 in F. pseudograminearum. B: Protein domains of FpMcas. C: Phylogenetic tree of FpMcas. D: Sequence analysis of the P20 subunit
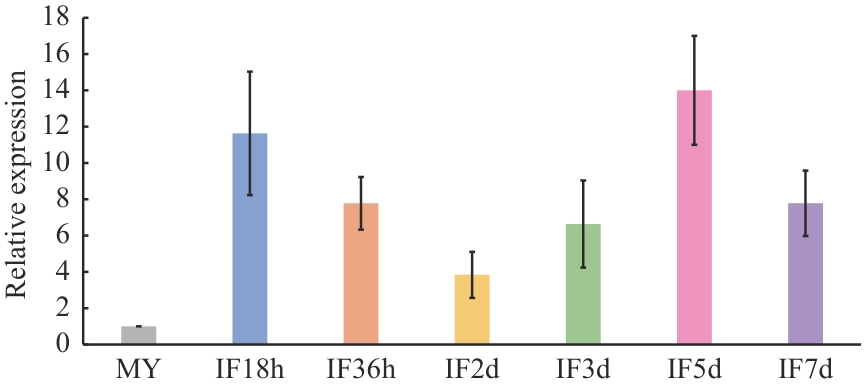
Fig. 2 Expression analysis of FpMca3 during the infection stages of F. pseudograminearumThe expression of FpMca3 in mycelia was arbitrarily set as 1. Bars denote standard errors from four repeated experiments. IF18h, IF36h, IF2d, IF3d, IF5d, and IF7d represent 18 h, 36 h, 2 d, 3 d, 5 d, and 7 d after inoculation of wheat (Aikang 58) coleoptiles with the wild-type mycelium of F. pseudograminearum, respectively
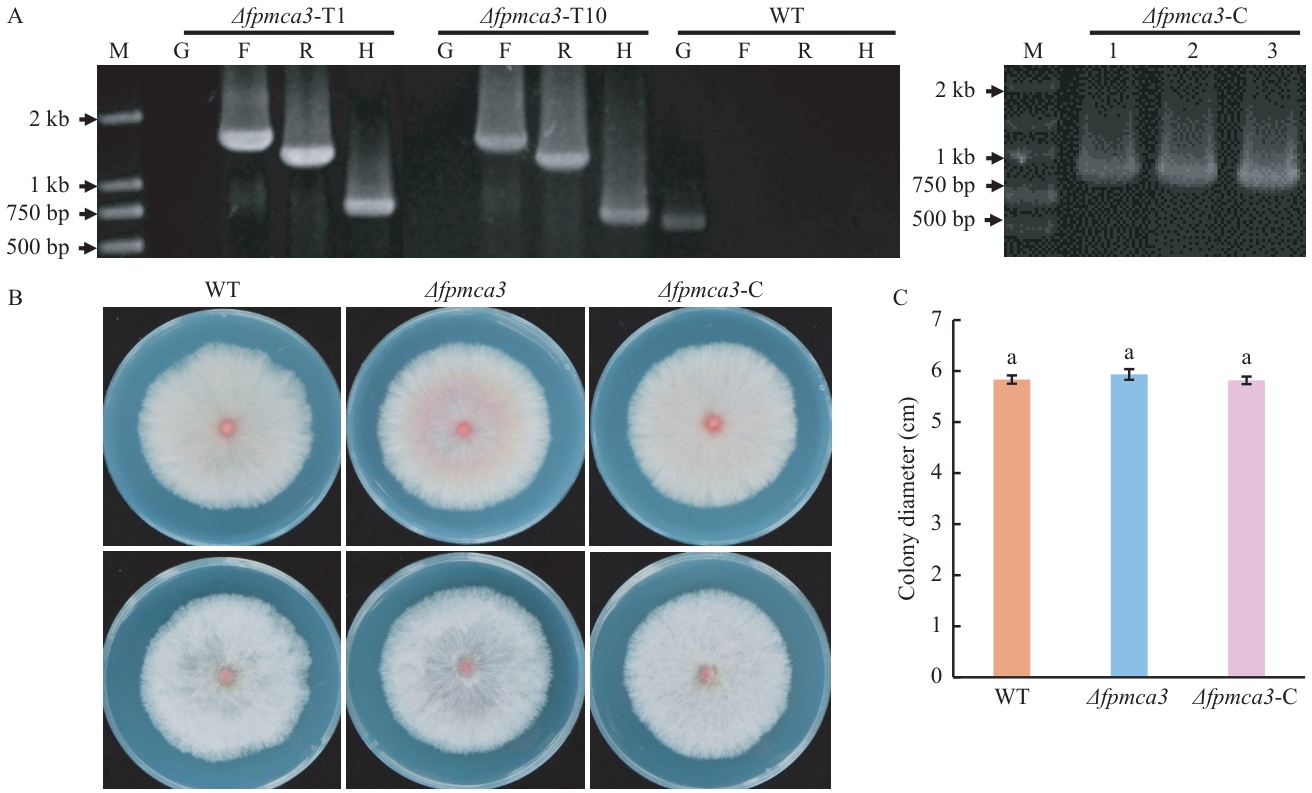
Fig. 3 Generation and growth phenotype determination of FpMca3-knockout mutantA: PCR verification of the FpMca3-knockout mutants and FpMca3-complemented strains. B: Growth assay of the FpMca3-knockout mutant. The upper row shows the reverse side of the colonies, while the lower row displays the front side. C: Measurement of colony diameter. Lowercase letters indicate data difference at the P<0.05 level
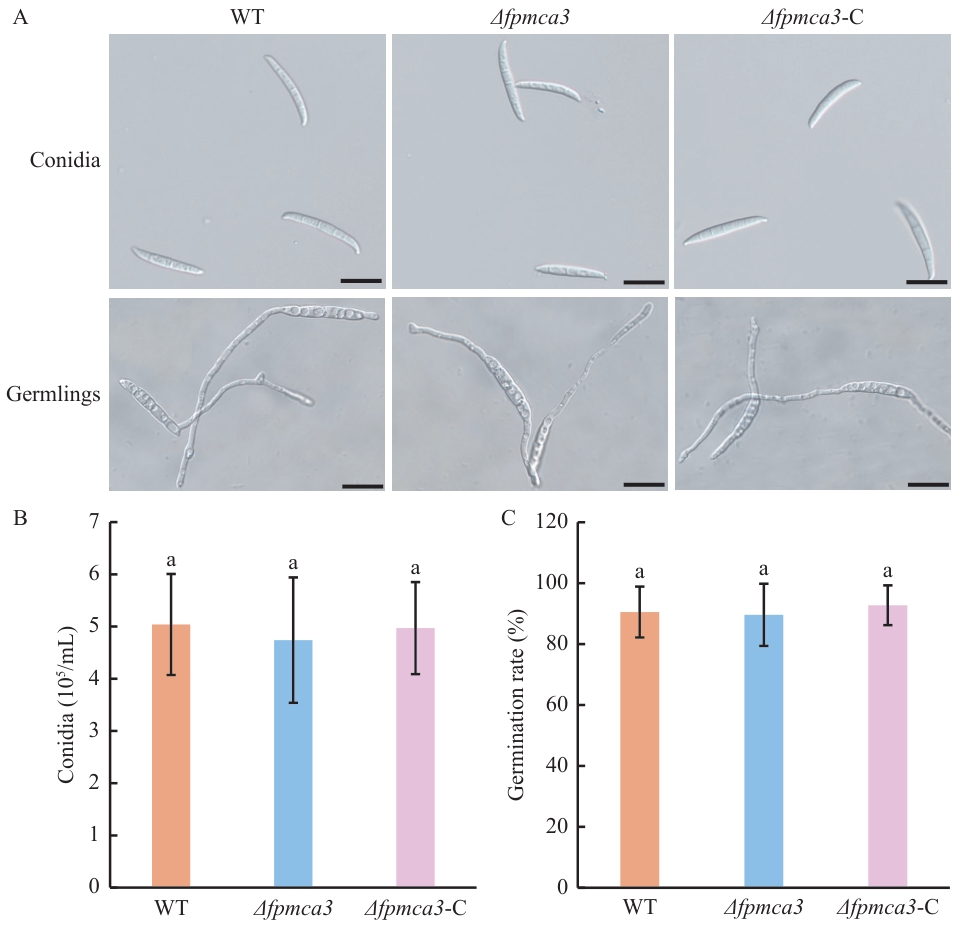
Fig. 4 Conidial production and germination assaysA: Morphology of conidia and germination of the wild-type, Δfpmca3 and Δfpmca3-C F. pseudograminearum. B: The conidia quantity of the wild-type, Δfpmca3 and Δfpmca3-C strains. C: Germination rates of the wild-type, Δfpmca3 and Δfpmca3-C strains. Lowercase letters indicate data difference at the P<0.05 level
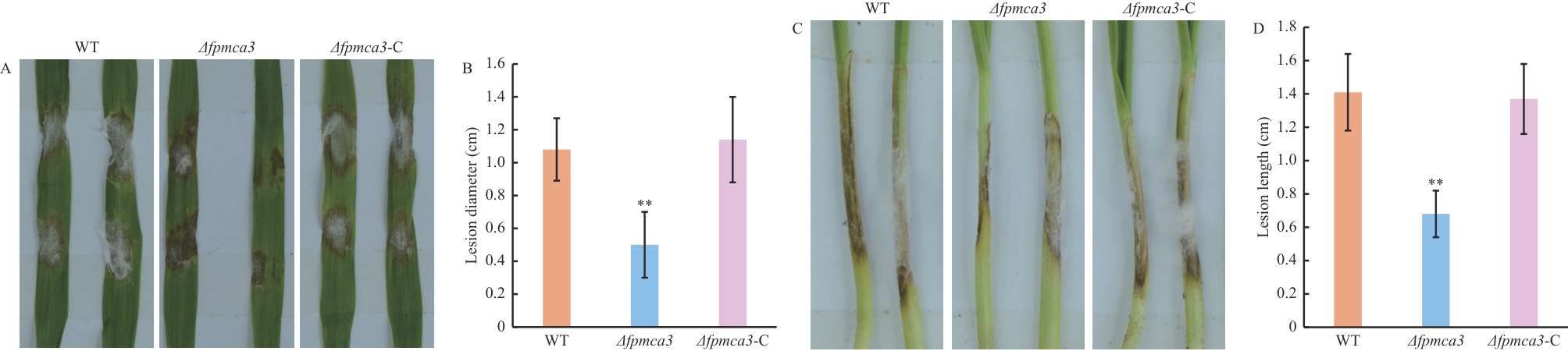
Fig. 5 Pathogenicity assay of F. pseudograminearum on barley leaves and wheat coleoptilesA: Phenotypes of lesions on barley leaves inoculated with the wild-type, Δfpmca3 and Δfpmca3-C F. pseudograminearum. B: Diameters of lesions on barley leaves measured at 4 d after inoculation. C: Phenotypes of lesions on wheat coleoptiles inoculated with the wild-type, Δfpmca3 and Δfpmca3-C strains. D: Diameters of lesions on wheat coleoptiles measured at 4 d after inoculation. ** P<0.01

Fig. 6 Pathogenicity assay of F. pseudograminearum in pot trialsA: Wheat growth induced by F. pseudograminearum infection. B: Crown rot disease index (DSI) in wheat seedlings infected with different F. pseudograminearum strains. ** P<0.01
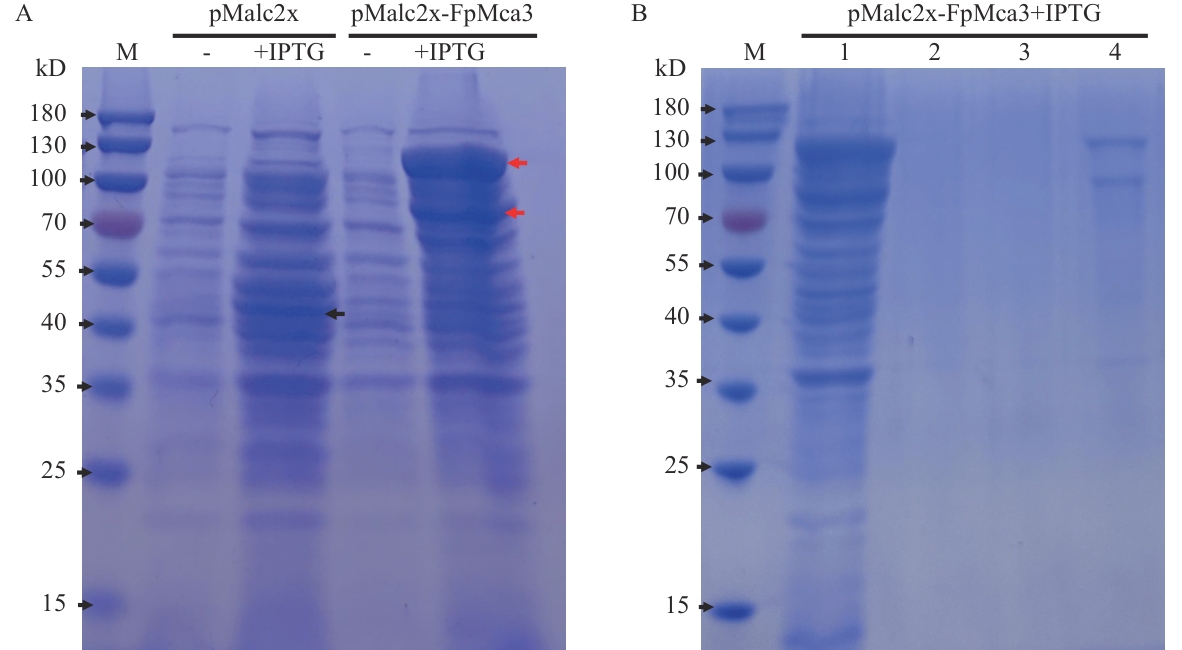
Fig. 7 Detection of prokaryotically expressed FpMca3 proteinA: IPTG-induced expression of FpMca3 in prokaryotic cells. The black arrow marks the position of MBP protein expressed by the pMalc2x empty vector control. The red arrows mark the FpMca3-MBP fusion protein expressed in the recombinant strain. B: Purification of FpMca3-MBP. 1: Unpurified protein. 2: Supernatant (post-binding, CK1). 3: Supernatant after washing with MBP buffer (CK2). 4: Protein after eluted with PBS
| [1] | Choi CJ, Berges JA. New types of metacaspases in phytoplankton reveal diverse origins of cell death proteases [J]. Cell Death Dis, 2013, 4(2): e490. |
| [2] | Fagundes D, Bohn B, Cabreira C, et al. Caspases in plants: metacaspase gene family in plant stress responses [J]. Funct Integr Genomics, 2015, 15(6): 639-649. |
| [3] | 姚绍嫦, 潘东进, 高程海. 海洋浮游植物metacaspase蛋白酶研究进展 [J]. 海洋科学, 2019, 43(8): 108-116. |
| Yao SC, Pan DJ, Gao CH. Research progress of metacaspase proteases in marine phytoplankton [J]. Mar Sci, 2019, 43(8): 108-116. | |
| [4] | Tsiatsiani L, Van Breusegem F, Gallois P, et al. Metacaspases [J]. Cell Death Differ, 2011, 18(8): 1279-1288. |
| [5] | Ghorbani N, Yaghubi R, Davoodi J, et al. How does caspases regulation play role in cell decisions? apoptosis and beyond [J]. Mol Cell Biochem, 2024, 479(7): 1599-1613. |
| [6] | Hill SM, Nyström T. The dual role of a yeast metacaspase: What doesn't kill you makes you stronger [J]. Bioessays, 2015, 37(5): 525-531. |
| [7] | Hill SM, Hao XX, Liu BD, et al. Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae [J]. Science, 2014, 344(6190): 1389-1392. |
| [8] | Mukherjee D, Gupta S, Saran N, et al. Induction of apoptosis-like cell death and clearance of stress-induced intracellular protein aggregates: dual roles for Ustilago maydis metacaspase Mca1 [J]. Mol Microbiol, 2017, 106(5): 815-831. |
| [9] | Coll NS, Vercammen D, Smidler A, et al. Arabidopsis type I metacaspases control cell death [J]. Science, 2010, 330(6009): 1393-1397. |
| [10] | Ruiz-Solaní N, Salguero-Linares J, Armengot L, et al. Arabidopsis metacaspase MC1 localizes in stress granules, clears protein aggregates, and delays senescence [J]. Plant Cell, 2023, 35(9): 3325-3344. |
| [11] | Pitsili E, Rodriguez-Trevino R, Ruiz-Solani N, et al. A phloem-localized Arabidopsis metacaspase (AtMC3) improves drought tolerance [J]. New Phytol, 2023, 239(4): 1281-1299. |
| [12] | Hander T, Fernández-Fernández ÁD, Kumpf RP, et al. Damage on plants activates Ca2+-dependent metacaspases for release of immunomodulatory peptides [J]. Science, 2019, 363(6433): eaar7486. |
| [13] | Zhu P, Yu XH, Wang C, et al. Structural basis for Ca2+-dependent activation of a plant metacaspase [J]. Nat Commun, 2020, 11(1): 2249. |
| [14] | Madeo F, Herker E, Maldener C, et al. A caspase-related protease regulates apoptosis in yeast [J]. Mol Cell, 2002, 9(4): 911-917. |
| [15] | Shrestha A, Brunette S, Stanford WL, et al. The metacaspase Yca1 maintains proteostasis through multiple interactions with the ubiquitin system [J]. Cell Discov, 2019, 5: 6. |
| [16] | Eyssen LE, Coetzer THT. Expression, purification and characterisation of Trypanosoma congolense metacaspase 5 (TcoMCA5) - a potential drug target for animal African trypanosomiasis [J]. Protein Expr Purif, 2019, 164: 105465. |
| [17] | Kumar B, Mohammad T, Amaduddin, et al. Targeting metacaspase-3 from Plasmodium falciparum towards antimalarial therapy: a combined approach of in-silico and in-vitro investigation [J]. J Biomol Struct Dyn, 2021, 39(2): 421-430. |
| [18] | Fernandez J, Lopez V, Kinch L, et al. Role of two metacaspases in development and pathogenicity of the rice blast fungus Magnaporthe oryzae [J]. mBio, 2021, 12(1): e03471-20. |
| [19] | Zhou HF, He XL, Wang S, et al. Diversity of the Fusarium pathogens associated with crown rot in the Huanghuai wheat-growing region of China [J]. Environ Microbiol, 2019, 21(8): 2740-2754. |
| [20] | 赵静雅, 凡卓, 彭梦雅, 等. FpStuA参与假禾谷镰孢的产孢和致病性 [J]. 植物病理学报, 2022, 52(1): 37-46. |
| Zhao JY, Fan Z, Peng MY, et al. FpStuA is involved in conidiation and pathogenicity of Fusarium pseudograminearum [J]. Acta Phytopathol Sin, 2022, 52(1): 37-46. | |
| [21] | Wang LM, Zhang YF, Du ZL, et al. FpPDE1 function of Fsarium pseudograminearum on pathogenesis in wheat [J]. J Integr Agric, 2017, 16(11): 2504-2512. |
| [22] | Chen LL, Geng XJ, Ma YM, et al. Identification of basic helix-loop-helix transcription factors reveals candidate genes involved in pathogenicity of Fusarium pseudograminearum [J]. Can J Plant Pathol, 2019, 41(2): 200-208. |
| [23] | Yang X, Pan YB, Singh PK, et al. Investigation and genome-wide association study for Fusarium crown rot resistance in Chinese common wheat [J]. BMC Plant Biol, 2019, 19(1): 153. |
| [24] | Chen LL, Ma YM, Peng MY, et al. Analysis of apoptosis-related genes reveals that apoptosis functions in conidiation and pathogenesis of Fusarium pseudograminearum [J]. mSphere, 2021, 6(1): e01140-20. |
| [25] | Murray GM, Brennan JP. Estimating disease losses to the Australian wheat industry [J]. Australas Plant Pathol, 2009, 38(6): 558-570. |
| [26] | Bragard C, Baptista P, Chatzivassiliou E, et al. Pest categorisation of Fusarium pseudograminearum [J]. EFSA J, 2022, 20(6): e07399. |
| [27] | Kazan K, Gardiner DM. Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: recent progress and future prospects [J]. Mol Plant Pathol, 2018, 19(7): 1547-1562. |
| [28] | Li HL, Yuan HX, Fu B, et al. First report of Fusarium pseudograminearum causing crown rot of wheat in Henan, China [J]. Plant Dis, 2012, 96(7): 1065. |
| [29] | 栾冬冬, 贾吉玉, 王光州, 等. 中国小麦茎基腐病的发生现状及防治策略[J]. 麦类作物学报 2022, 42(4): 512-520. |
| Luan DD, Jia JY, Wang GZ, et al. Occurrence status and control strategies of wheat crown rot in China [J]. J Triticeae Crops, 2022, 42(4): 512-520. | |
| [30] | 宋嘉庆, 潘鑫, 闫书味, 等. 假禾谷镰孢菌的分离及其对小麦茎基部和穗部的致病力分析 [J]. 麦类作物学报, 2022, 42(12): 1575-1581. |
| Song JQ, Pan X, Yan SW, et al. Analysis of the pathogenicity of Fusarium pseudograminearum on wheat stem bases and panicles [J]. J Triticeae Crops, 2022, 42(12): 1575-1581. | |
| [31] | Aoki T, O'Donnell K. Morphological and molecular characterization of Fusarium pseudograminearum sp. nov., formerly recognized as the group 1 population of F. graminearum [J]. Mycologia, 1999, 91(4): 597-609. |
| [32] | Tunali B, Obanor F, Erginbaş G, et al. Fitness of three Fusarium pathogens of wheat [J]. FEMS Microbiol Ecol, 2012, 81(3): 596-609. |
| [33] | Powell JJ, Carere J, Fitzgerald TL, et al. The Fusarium crown rot pathogen Fusarium pseudograminearum triggers a suite of transcriptional and metabolic changes in bread wheat (Triticum aestivum L.) [J]. Ann Bot, 2017, 119(5): 853-867. |
| [34] | Gardiner DM, McDonald MC, Covarelli L, et al. Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts [J]. PLoS Pathog, 2012, 8(9): e1002952. |
| [1] | CHAI Jun-fa, HONG Bo, JIA Yan-xia. Combined Transcriptomic and Metabolomic Analysis of Virulence Differences among Three Lecanicillium lecanii Strains [J]. Biotechnology Bulletin, 2025, 41(8): 311-321. |
| [2] | TIAN Tong-tong, GE Jia-zhen, GAO Peng-cheng, LI Xue-rui, SONG Guo-dong, ZHENG Fu-ying, CHU Yue-feng. Whole Genome Sequencing and Bioinformatics Analysis of Mycoplasma ovipneumoniae GH3-3 Strain [J]. Biotechnology Bulletin, 2024, 40(7): 323-334. |
| [3] | WANG Zi, SHI Jin-chuan, WANG Yong-qiang, SUN Miao, MENG Ling-hao, GENG Chao, LIU Kai. Whole Genome Sequencing and Genome Evolution Analysis of Capsular Serotype A and D Pasteurella multocida of Bovine [J]. Biotechnology Bulletin, 2024, 40(12): 282-290. |
| [4] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [5] | CHEN Bao-qiang, LI Ying-ying, MA Bo-ya, ROUZHAGULI Malike, YOULITUZI Naibi, SONG Jin-di, LIU Jun, WANG Xi-dong. Functional Analysis of the Type III Secreted Effector Gene aop2 in Acidovorax citrulli [J]. Biotechnology Bulletin, 2023, 39(6): 286-297. |
| [6] | HOU Xiao-yuan, CHE Zheng-zheng, LI Heng-jing, DU Chong-yu, XU Qian, WANG Qun-qing. Construction of the Soybean Membrane System cDNA Library and Interaction Proteins Screening for Effector PsAvr3a [J]. Biotechnology Bulletin, 2023, 39(4): 268-276. |
| [7] | WU Li-dan, RAN Xue-qin, NIU Xi, HUANG Shi-hui, LI Sheng, WANG Jia-fu. Genome Comparison and Virulence Factor Analysis of Pathogenic Escherichia coli from Porcine [J]. Biotechnology Bulletin, 2023, 39(12): 287-299. |
| [8] | LIU Li-hui, CHU Jin-hua, SUI Yu-xin, CHEN Yang, CHENG Gu-yue. Research Progress of Main Virulence Factors in Salmonella [J]. Biotechnology Bulletin, 2022, 38(9): 72-83. |
| [9] | ZHAO Jing-ya, PENG Meng-ya, ZHANG Shi-yu, SHAN Yi-xuan, XING Xiao-ping, SHI Yan, LI Hai-yang, YANG Xue, LI Hong-lian, CHEN Lin-lin. Role of C2H2 Zinc Finger Transcription Factor FpCzf7 in the Growth and Pathogenicity of Fusarium pseudograminearum [J]. Biotechnology Bulletin, 2022, 38(8): 216-224. |
| [10] | CHEN Fu-nuan, HUANG Yu, CAI Jia, WANG Zhong-liang, JIAN Ji-chang, WANG Bei. Structure of ABC Transporter and Research Progress of It in Bacterial Pathogenicity [J]. Biotechnology Bulletin, 2022, 38(6): 43-52. |
| [11] | TIAN Li, LI Jun-jiao, DAI Xiao-feng, ZHANG Dan-dan, CHEN Jie-yin. From Functional Genes to Biological Characteristics:The Molecular Basis of Pathogenicity in Verticillium dahliae [J]. Biotechnology Bulletin, 2022, 38(1): 51-69. |
| [12] | LIU Hui, DENG Zhi, YANG Hong, DAI Long-jun, LI De-jun. Expression and Stress Tolerance Analysis of HbMC2 Gene from Hevea brasliensis in Yeast [J]. Biotechnology Bulletin, 2018, 34(9): 202-208. |
| [13] | WANG Hong-yang, QIN Li-juan, TANG Wei, TIAN Zhen-dong. Research Advances on Phytophthora infestans RXLR Effector Proteins [J]. Biotechnology Bulletin, 2018, 34(2): 102-111. |
| [14] | YAN Yu-ping, ZHONG Xi, WANG Xue-feng. Research Progress on Small Non-coding RNA of the Xanthomonas spp. [J]. Biotechnology Bulletin, 2017, 33(3): 22-28. |
| [15] | Ling Kong,Ding Shihua, Jin Juan,Wu Xingzhen. Detection of Pathogenic Aeromonas hydrophila in Andrias davidianus by Quadruple PCR [J]. Biotechnology Bulletin, 2014, 30(9): 201-207. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||