








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (5): 273-280.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1181
Previous Articles Next Articles
QU Huan1( ), LI Cheng1, CHEN Rui1, LIAO Yi-jie1, CAO San-jie1,2,3, WEN Yi-ping1, YAN Qi-gui1, HUANG Xiao-bo1,2,3(
), LI Cheng1, CHEN Rui1, LIAO Yi-jie1, CAO San-jie1,2,3, WEN Yi-ping1, YAN Qi-gui1, HUANG Xiao-bo1,2,3( )
)
Received:2020-09-18
Online:2021-05-26
Published:2021-06-11
Contact:
HUANG Xiao-bo
E-mail:1157759897@qq.com;rsghb110@126.com
QU Huan, LI Cheng, CHEN Rui, LIAO Yi-jie, CAO San-jie, WEN Yi-ping, YAN Qi-gui, HUANG Xiao-bo. Truncated Expression of the S1-CTD Fragment of Porcine Deltacoronavirus and Establishment of an Indirect ELISA for Detecting Its Antibody[J]. Biotechnology Bulletin, 2021, 37(5): 273-280.
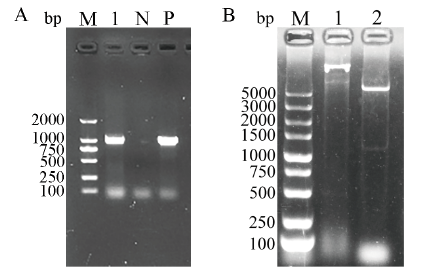
Fig. 2 Identification of recombinant plasmid pET28a-S1CTD by PCR (A) and double digestion (B) A: Identification of recombinant plasmid pET28a-S1CTD by PCR (M: DL2000 DNA marker, 1: Target gene, N: Negative control, P: Positive control). B: Identification of recombinant plasmid pET28a-S1CTD by double digestion (M: DL5000 DNA marke, 1: Undigested recombinant plasmid, 2: Digested recombinant plasmid)

Fig. 3 The SDS-PAGE and Western blot analysis of recombinant S1-CTD protein A: The SDS-PAGE analysis of S1-CTD (M: Protein marker, 1: Post-induced BL21-pET28a, 2: Pre-induced pET28a-S1-CTD, 3: Post-induced pET28a-S1-CTD, 4: Hyperacoustic full bacteria after induction, 5: Hyperacoustic supernatant, 6: Hyperacoustic precipitation). B: Purification of S1-CTD protein (M: Protein marker, 1: Pre-lysing inclusion body protein, 2: Dissolved inclusion body protein, 3: Target protein eluted by 50 mmol/L imidazole, 4: Target protein eluted by 150 mmol/L imidazole, 5: Target protein eluted by 250 mmol/L imidazole). C: The western blot analysis of S1-CTD (M: Protein marker, 1-4: BL21-pET28a, pET28a-S1-CTD, pET-28a-E, pET-28a-N proteins reacted with pig anti-PDCoV positive serum, 5: pET28a-S1-CTD protein reacted with pig anti-PDCoV negative serum)
| 血清编号 Serum No. | 批内变异系数 Intra batch CV % | 批间变异系数 Inter batch CV % | ||
|---|---|---|---|---|
| ±SD | 变异系数 CV/% | ±SD | 变异系数 CV/% | |
| 1 | 0.136±0.011 | 8.07 | 0.190±0.017 | 8.85 |
| 2 | 0.110±0.011 | 9.57 | 0.217±0.020 | 9.22 |
| 3 | 0.127±0.004 | 3.48 | 0.122±0.012 | 9.98 |
| 4 | 0.124±0.005 | 4.35 | 0.155±0.013 | 8.29 |
| 5 | 0.113±0.003 | 2.65 | 0.909±0.055 | 6.01 |
| 6 | 1.251±0.043 | 3.47 | 1.022±0.079 | 7.75 |
| 7 | 0.946±0.032 | 3.43 | 0.928±0.085 | 9.12 |
Table 1 Repeatability of the indirect ELISA
| 血清编号 Serum No. | 批内变异系数 Intra batch CV % | 批间变异系数 Inter batch CV % | ||
|---|---|---|---|---|
| ±SD | 变异系数 CV/% | ±SD | 变异系数 CV/% | |
| 1 | 0.136±0.011 | 8.07 | 0.190±0.017 | 8.85 |
| 2 | 0.110±0.011 | 9.57 | 0.217±0.020 | 9.22 |
| 3 | 0.127±0.004 | 3.48 | 0.122±0.012 | 9.98 |
| 4 | 0.124±0.005 | 4.35 | 0.155±0.013 | 8.29 |
| 5 | 0.113±0.003 | 2.65 | 0.909±0.055 | 6.01 |
| 6 | 1.251±0.043 | 3.47 | 1.022±0.079 | 7.75 |
| 7 | 0.946±0.032 | 3.43 | 0.928±0.085 | 9.12 |
| 检测方法 Detection method | VNT | |||
|---|---|---|---|---|
| 阳性数 Positive number | 阴性数 Negative number | 合计 Total | ||
| 阳性数 Positive number | 9 | 3 | 12 | |
| 阴性数 Negative number | 2 | 16 | 18 | |
| 间接ELISA | 合计 Total | 11 | 19 | 30 |
| 阳性符合率 Positive coincidence rate | 81.8% | |||
| Indirect ELISA | 阴性符合率 Negative coincidence rate | 84.2% | ||
| 总符合率 Total coincidence rate | 83.3% | |||
Table 2 Comparative detection of serum samples by indirect ELISA and VNT
| 检测方法 Detection method | VNT | |||
|---|---|---|---|---|
| 阳性数 Positive number | 阴性数 Negative number | 合计 Total | ||
| 阳性数 Positive number | 9 | 3 | 12 | |
| 阴性数 Negative number | 2 | 16 | 18 | |
| 间接ELISA | 合计 Total | 11 | 19 | 30 |
| 阳性符合率 Positive coincidence rate | 81.8% | |||
| Indirect ELISA | 阴性符合率 Negative coincidence rate | 84.2% | ||
| 总符合率 Total coincidence rate | 83.3% | |||
| 来源 Origin | 样本数 Sample number | 阳性总数 Positive number | 阳性率 Positive rate/% |
|---|---|---|---|
| 成都 Chengdu | 52 | 34 | 65.38 |
| 南充 Nanchong | 57 | 26 | 45.61 |
| 绵阳 Mianyang | 30 | 12 | 40 |
| 眉山 Meishan | 33 | 3 | 9.09 |
| 遂宁 Suining | 16 | 0 | 0 |
| 德阳 Deyang | 61 | 24 | 39.34 |
| 达州 Dazhou | 27 | 27 | 100 |
| 泸州 Luzhou | 9 | 8 | 88.89 |
| 宜宾 Yibin | 1 | 1 | 100 |
| 阿坝 Aba | 23 | 22 | 95.65 |
| 巴中 Bazhong | 90 | 84 | 93.33 |
| 凉山 Liangshan | 4 | 0 | 0 |
| 资阳 Ziyang | 1 | 0 | 0 |
| 雅安 Ya’an | 56 | 0 | 0 |
| 内江 Neijiang | 12 | 0 | 0 |
| 乐山 Leshan | 18 | 0 | 0 |
| 总数 Total | 490 | 241 | 49.18 |
Table 3 Clinical detection of PDCoV samples
| 来源 Origin | 样本数 Sample number | 阳性总数 Positive number | 阳性率 Positive rate/% |
|---|---|---|---|
| 成都 Chengdu | 52 | 34 | 65.38 |
| 南充 Nanchong | 57 | 26 | 45.61 |
| 绵阳 Mianyang | 30 | 12 | 40 |
| 眉山 Meishan | 33 | 3 | 9.09 |
| 遂宁 Suining | 16 | 0 | 0 |
| 德阳 Deyang | 61 | 24 | 39.34 |
| 达州 Dazhou | 27 | 27 | 100 |
| 泸州 Luzhou | 9 | 8 | 88.89 |
| 宜宾 Yibin | 1 | 1 | 100 |
| 阿坝 Aba | 23 | 22 | 95.65 |
| 巴中 Bazhong | 90 | 84 | 93.33 |
| 凉山 Liangshan | 4 | 0 | 0 |
| 资阳 Ziyang | 1 | 0 | 0 |
| 雅安 Ya’an | 56 | 0 | 0 |
| 内江 Neijiang | 12 | 0 | 0 |
| 乐山 Leshan | 18 | 0 | 0 |
| 总数 Total | 490 | 241 | 49.18 |
| [1] | Li G, Chen Q, Harmon KM, et al. Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734[J]. Genome Announc, 2014,2(2):e00278-14. |
| [2] |
Woo PC, Lau SK, Lam CS, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus[J]. J Virol, 2012,86(7):3995-4008.
doi: 10.1128/JVI.06540-11 URL |
| [3] | Marthaler D, Jiang Y, Collins J, et al. Complete genome sequence of strain SDCV/USA/Illinois121/2014, a porcine deltacoronavirus from the United States[J]. Genome Announc, 2014,2(2):e00218-14. |
| [4] |
Wang L, Byrum B, Zhang Y. Porcine coronavirus HKU15 detected in 9 US states, 2014[J]. Emerg Infect Dis, 2014,20(9):1594-1595.
doi: 10.3201/eid2009.140756 URL |
| [5] |
Niederwerder MC, Hesse RA. Swine enteric coronavirus disease:A review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada[J]. Transbound Emerg Dis, 2018,65(3):660-675.
doi: 10.1111/tbed.12823 pmid: 29392870 |
| [6] |
Mai K, Feng J, Chen G, et al. The detection and phylogenetic analysis of porcine deltacoronavirus from Guangdong province in southern China[J]. Transbound Emerg Dis, 2018,65(1):166-173.
doi: 10.1111/tbed.12644 pmid: 28345292 |
| [7] |
Jang G, Lee KK, Kim SH, et al. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014-2016[J]. Transbound Emerg Dis, 2017,64(5):1364-1370.
doi: 10.1111/tbed.12690 pmid: 28758347 |
| [8] |
Saeng-Chuto K, Lorsirigool A, Temeeyasen G, et al. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015[J]. Transbound Emerg Dis, 2017,64(1):3-10.
doi: 10.1111/tbed.12585 pmid: 27718337 |
| [9] |
Suzuki T, Shibahara T, Imai N, et al. Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus[J]. Infect Genet Evol, 2018,61:176-182.
doi: 10.1016/j.meegid.2018.03.030 URL |
| [10] |
Dong N, Fang L, Zeng S, et al. Porcine deltacoronavirus in mainland China[J]. Emerg Infect Dis, 2015,21(12):2254-2255.
doi: 10.3201/eid2112.150283 URL |
| [11] |
Wang M, Wang Y, Baloch AR, et al. Detection and genetic characterization of porcine deltacoronavirus in Tibetan pigs surrounding the Qinghai-Tibet Plateau of China[J]. Transbound Emerg Dis, 2018,65(2):363-369.
doi: 10.1111/tbed.12819 pmid: 29363281 |
| [12] |
Zhang HL, Liang QQ, Li BX, et al. Prevalence, phylogenetic and evolutionary analysis of porcine deltacoronavirus in Henan province, China[J]. Prev Vet Med, 2019,166:8-15.
doi: 10.1016/j.prevetmed.2019.02.017 URL |
| [13] | Lee S, Lee C. Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014[J]. Genome Announc, 2014,2(6):e01191-14. |
| [14] | 方谱县, 方六荣, 董楠, 等. 猪δ冠状病毒的研究进展[J]. 病毒学报, 2016,32(2):243-248. |
| Fang PX, Fang LR, Dong N, et al. Research advances in the porcine deltacoronavirus[J]. Chinese Journal of Virology, 2016,32(2):243-248. | |
| [15] | Shang J, Zheng Y, Yang Y, et al. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state[J]. J Virol, 2018,92(4):e01556-17. |
| [16] |
Chen R, Fu J, Hu J, et al. Identification of the immunodominant neutralizing regions in the spike glycoprotein of porcine deltacoronavirus[J]. Virus Res, 2020,276:197834.
doi: 10.1016/j.virusres.2019.197834 URL |
| [17] |
Brian DA, Baric RS. Coronavirus genome structure and replication[J]. Curr Top Microbiol Immunol, 2005,287:1-30.
pmid: 15609507 |
| [18] |
Sun DB, Feng L, Shi HY, et al. Spike protein region(aa 636789)of porcine epidemic diarrhea virus is essential for induction of neutralizing antibodies[J]. Acta Virol, 2007,51(3):149-156.
pmid: 18076304 |
| [19] |
Carvajal A, Lanza I, Diego R, et al. Evaluation of a blocking ELISA using monoclonal antibodies for the detection of porcine epidemic diarrhea virus and its antibodies[J]. J Vet Diagn Invest, 1995,7(1):60-64.
pmid: 7779966 |
| [20] |
Su M, Li C, Guo D, et al. A recombinant nucleocapsid protein-based indirect enzyme-linked immunosorbent assay to detect antibodies against porcine deltacoronavirus[J]. J Vet Med Sci, 2016,78(4):601-606.
doi: 10.1292/jvms.15-0533 URL |
| [21] | 杨浩, 方六荣, 董楠, 等. 基于猪德尔塔冠状病毒重组核衣壳蛋白的 ELISA抗体检测方法的建立与评价[J]. 微生物学通报, 2017,44(12):2830-2838. |
| Yang H, Fang LR, Dong N, et al. Development and evaluation of an indirect ELISA based on recombinant nucleocapsid protein for detecting antibodies against porcine deltacoronavirus[J]. Microbiology China, 2017,44(12):2830-2838. | |
| [22] |
Luo SX, Fan JH, Opriessnig T, et al. Development and application of a recombinant M protein-based indirect ELISA for the detection of porcine deltacoronavirus IgG antibodies[J]. J Virol Methods, 2017,249:76-78.
doi: 10.1016/j.jviromet.2017.08.020 URL |
| [23] | 殷震. 动物病毒学基础[M]. 长春: 吉林人民出版社, 1980. |
| Yin Z. Fundamentals of animal virology[M]. Changchun: Jilin People’s Press, 1980. | |
| [24] | Wang L, Byrum B, Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014[J]. Emerg Infect Dis, 2014,20(7):1227-1230. |
| [25] | 孙东波. 猪流行性腹泻病毒S蛋白抗原表位鉴定及受体结合域的初步筛选[D]. 北京:中国农业科学院, 2008. |
| Sun DB. Identification of antigenic epitopes and screening receptor binding domain on the S protein of porcine epidemic diarrhea virus[D]. Beijing:Chinese Academy of Agricultural Sciences, 2008. | |
| [26] |
Ma Y, Zhang Y, Liang X, et al. Two-way antigenic cross-reactivity between porcine epidemic diarrhea virus and porcine deltacoronavirus[J]. Vet Microbiol, 2016,186:90-96.
doi: 10.1016/j.vetmic.2016.02.004 URL |
| [27] |
Gimenez-Lirola LG, Zhang J, Carrillo-Avila JA, et al. Reactivity of porcine epidemic diarrhea virus structural proteins to antibodies against porcine enteric coronaviruses:diagnostic implications[J]. J Clin Microbiol, 2017,55(5):1426-1436.
doi: 10.1128/JCM.02507-16 URL |
| [28] |
Thachil A, Gerber PF, Xiao CT, et al. Development and application of an ELISA for the detection of porcine deltacoronavirus IgG antibodies[J]. PLoS One, 2015,10(4):e0124363.
doi: 10.1371/journal.pone.0124363 URL |
| [29] |
Sestak K, Zhou Z, Shoup DI, et al. Evaluation of the baculovirus-expressed S glycoprotein of transmissible gastroenteritis virus(TGEV)as antigen in a competition ELISA to differentiate porcine respiratory coronavirus from TGEV antibodies in pigs[J]. J Vet Diagn Invest, 1999,11(3):205-214.
pmid: 10353350 |
| [30] |
Gerber PF, Gong Q, Huang YW, et al. Detection of antibodies against porcine epidemic diarrhea virus in serum and colostrum by indirect ELISA[J]. Vet J, 2014,202(1):33-36.
doi: 10.1016/j.tvjl.2014.07.018 URL |
| [31] | 王经纬, 雷喜梅, 覃盼, 等. 猪丁型冠状病毒荧光定量 RT-PCR 与 S1 蛋白间接ELISA 检测方法的建立及应用[J]. 生物工程学报, 2017,33(8):1265-1275. |
| Wang JW, Lei XM, Qin P, et al. Development and application of real-time RT-PCR and S1 protein-based indirect ELISA for porcine deltacoronavirus[J]. Chin J Biotech, 2017,33(8):1265-1275. | |
| [32] |
Singh SM, Panda AK. Solubilization and refolding of bacterial inclusion body proteins[J]. J Biosci Bioeng, 2005,99(4):303-310.
doi: 10.1263/jbb.99.303 URL |
| [33] | Burgess RR. Refolding solubilized inclusion body proteins[J]. Methods Enzymol, 2009,463:259-282. |
| [34] | 逄凤娇, 俞正玉, 何孔旺, 等. 猪丁型冠状病毒重组N蛋白间接ELISA抗体检测方法的建立[J]. 中国预防兽医学报, 2017,39(6):461-465. |
| Pang FJ, Yu ZY, He KW, et al. Development of an indirect ELISA for detection antibody to porcine deltacoronavirus using recombinant N protein[J]. Chinese Journal of Preventive Veterinary Medicine, 2017,39(6):461-465. |
| [1] | MEI Huan, LI Yue, LIU Ke-meng, LIU Ji-hua. Study on the Biosynthesis of l-SLR by Efficient Prokaryotic Expression of Berberine Bridge Enzyme [J]. Biotechnology Bulletin, 2023, 39(7): 277-287. |
| [2] | SUO Qing-qing, WU Nan, YANG Hui, LI Li, WANG Xi-feng. Prokaryotic Expression,Antibody Preparation and Application of Rice Caffeoyl Coenzyme A-O-methyltransferase Gene [J]. Biotechnology Bulletin, 2022, 38(8): 135-141. |
| [3] | QIN Xue-jing, WANG Yu-han, CAO Yi-bo, ZHANG Ling-yun. Prokaryotic Expression and Preparation of Polyclonal Antibody of PwHAP5 Gene in Picea wilsonii [J]. Biotechnology Bulletin, 2022, 38(8): 142-149. |
| [4] | WANG Guang-li, FAN Chan, WANG Hui, LU Hui-fang, XIA Ling-yin, HUANG Jian, MIN Xun. Prokaryotic Expression,Purification,Identification,and Polyclonal Antibody Preparation of Vibrio cholerae Hemolysin HlyA [J]. Biotechnology Bulletin, 2022, 38(7): 269-277. |
| [5] | WANG Qiao-ju, HU Yu-meng, WEN Ya-ya, SONG Li, MENG Chuang, PAN Zhi-ming, JIAO Xin-an. Expression and Activity Identification of SARS-CoV-2 S1 Protein [J]. Biotechnology Bulletin, 2022, 38(3): 157-163. |
| [6] | SHEN Jun-qiang, ZHANG Li-ping, YU Rui-ming, WANG Yong-lu, PAN Li, LIU Xia, LIU Xin-sheng. Porcine Kobuvirus Structural Proteins VP0 and VP1 Prokaryotic Expression and Establishment of Indirect ELISA Method [J]. Biotechnology Bulletin, 2022, 38(10): 243-253. |
| [7] | SHAN Cao-mei, YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang. Cloning,and Functional Analysis of Gene OsRAI1 Resistant to Hirschmanniella mucronate in Rice [J]. Biotechnology Bulletin, 2021, 37(7): 146-155. |
| [8] | ZENG Fu-yuan, SU Ze-hui, ZHOU Shi-hui, XIE Miao, PANG Huan-ying. Prokaryotic Expression of the PEPCK Protein of Vibrio alginolyticus and Identification of Its Acetylation and Succinylation [J]. Biotechnology Bulletin, 2021, 37(5): 84-91. |
| [9] | ZHANG Xi-xi, ZHANG Yi-qing, LI Yu-lin, HAN Xiao, WANG Guo-qiang, WANG Xiao-jun, WANG Xu-dong, WANG Yun-long. Prokaryotic Expression,Purification and Application of N Protein C-terminal Recombinant Protein in Novel Coronavirus(SARS-CoV-2) [J]. Biotechnology Bulletin, 2021, 37(5): 92-97. |
| [10] | BAI Fu-mei, LI Zhi-min, WANG Xiao-qin, HU Zi-wei, BAO Ling-ling, LI Zhi-min. Biochemical Characterization and Structural Analysis of N-acetylornithine Transaminase from Synechocystis sp. PCC6803 [J]. Biotechnology Bulletin, 2021, 37(5): 98-107. |
| [11] | PENG Li-zhong, ZHANG Peng, ZHOU Wen-wen, ZENG Xu-hui, ZHANG Xiao-ning. Preparation and Multi-purpose Validation of Sperm-specific Protein Cabs1 Polyclonal Antibody [J]. Biotechnology Bulletin, 2021, 37(3): 261-270. |
| [12] | HE Yang, YU Qiao-ling, WANG Jun, QIN Chuan-jie, LI Hua-tao. Advances in Prokaryotic Expression Gene of Tilapia [J]. Biotechnology Bulletin, 2021, 37(2): 195-202. |
| [13] | TANG Lu, DONG Li-ping, YIN Mo-li, LIU Lei, DONG Yuan, WANG Hui-yan. Preparation and Identification of a Novel FGF20 Monoclonal Antibody [J]. Biotechnology Bulletin, 2021, 37(10): 179-185. |
| [14] | DUAN Ying-ce, HU Zi-yi, YANG Fan, LI Jin-tao, WU Xiang-li, ZHANG Rui-ying. Cloning and Expression Analysis of Oxaloacetate Hydrolase(LeOAH1)Gene from Lentinula edodes [J]. Biotechnology Bulletin, 2020, 36(9): 227-234. |
| [15] | CHEN Rui, FU Jia-yu, LIU Hao-yu, LI Cheng, ZHAO Yu-jia, HU Jing-fei, QU Huan, CAO San-jie, WEN Xin-tian, WEN Yi-ping, ZHAO Qin, WU Rui, HUANG Xiao-bo. Prokaryotic Expression and Polyclonal Antibody Preparation of Porcine Deltacoronavirus(PDCoV)N Protein [J]. Biotechnology Bulletin, 2020, 36(8): 104-110. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||