








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (9): 161-170.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0047
Previous Articles Next Articles
LIU Sha-yu( ), CAO Jian, LI Meng, LIU Zhi-qiang, LI Xiao-yu(
), CAO Jian, LI Meng, LIU Zhi-qiang, LI Xiao-yu( )
)
Received:2021-01-11
Online:2021-09-26
Published:2021-10-25
Contact:
LI Xiao-yu
E-mail:1091293310@qq.com;hdlixiaoyu@126.com
LIU Sha-yu, CAO Jian, LI Meng, LIU Zhi-qiang, LI Xiao-yu. Biological Function of a Zn2Cys6 Transcription Factor CgAswA in Colletotrichum gloeosporioides[J]. Biotechnology Bulletin, 2021, 37(9): 161-170.

Fig. 1 Homologous recombination principle of the gene CgaswA Sur is the chlorimuron-ethyl resistance gene,and black triangles indicate the primers used to verify gene-knockout mutants
| Primer | Primer sequence(5'-3') |
|---|---|
| CgaswAupF | CGGAATTCTCAGAAGAGGCACCGCAACGCAT |
| CgaswAupR | CCCAAGCTTATCTCCTTCATCCGACACGACTG |
| CgaswAF | ATGAACGACAACATACTCTACC |
| CgaswAR | TCAAGTGCACAAGAAGTCGAGG |
| CgaswAdownF | GCTCTAGACGCTCAACCACCTCATCGCTACC |
| CgaswAdownR | CGGAATTCCCGTCCGTTGTCTTGACCTTTGC |
| CgaswAUU | CTCCTTCGCTCACTTGCTCGCTC |
| PI | CAGGGTTTTCCCAGTCACGACGTTG |
| PI1 | GTATGTTGTGTGGAATTGTGAGCGG |
| CgaswADD | GCACAATGGAGATCTTCGCCCTT |
| CgaswAhbF | CGGAATTCTGCTTTCGCTCCCTCTGTTTCTT |
| CgaswAhbR | GCTCTAGAGTCCGTTGTCTTGACCTTTGCTG |
Table 1 Primer and sequence
| Primer | Primer sequence(5'-3') |
|---|---|
| CgaswAupF | CGGAATTCTCAGAAGAGGCACCGCAACGCAT |
| CgaswAupR | CCCAAGCTTATCTCCTTCATCCGACACGACTG |
| CgaswAF | ATGAACGACAACATACTCTACC |
| CgaswAR | TCAAGTGCACAAGAAGTCGAGG |
| CgaswAdownF | GCTCTAGACGCTCAACCACCTCATCGCTACC |
| CgaswAdownR | CGGAATTCCCGTCCGTTGTCTTGACCTTTGC |
| CgaswAUU | CTCCTTCGCTCACTTGCTCGCTC |
| PI | CAGGGTTTTCCCAGTCACGACGTTG |
| PI1 | GTATGTTGTGTGGAATTGTGAGCGG |
| CgaswADD | GCACAATGGAGATCTTCGCCCTT |
| CgaswAhbF | CGGAATTCTGCTTTCGCTCCCTCTGTTTCTT |
| CgaswAhbR | GCTCTAGAGTCCGTTGTCTTGACCTTTGCTG |

Fig. 2 Protein domain and phylogenetic tree of CgAswA protein A:Protein domain analysis of CgAswA. B:Phylogenetic analysis of CgAswA. The sequence ID is in parentheses;the numbers on the horizontal lines represent evolutionary distances

Fig. 3 Verification of CgaswA gene knockout and com-plementation A:PCR results of CgaswAF / CgaswAR;B:PCR results of CgaswAUU / PI;C:PCR results of PI1 / CgaswADD. M:DL2 000 DNA marker;1:wild type;2:ΔCgaswA-3;3:ΔCgaswA-8;4:ΔCgaswA-11;5:ΔCgaswA/aswA
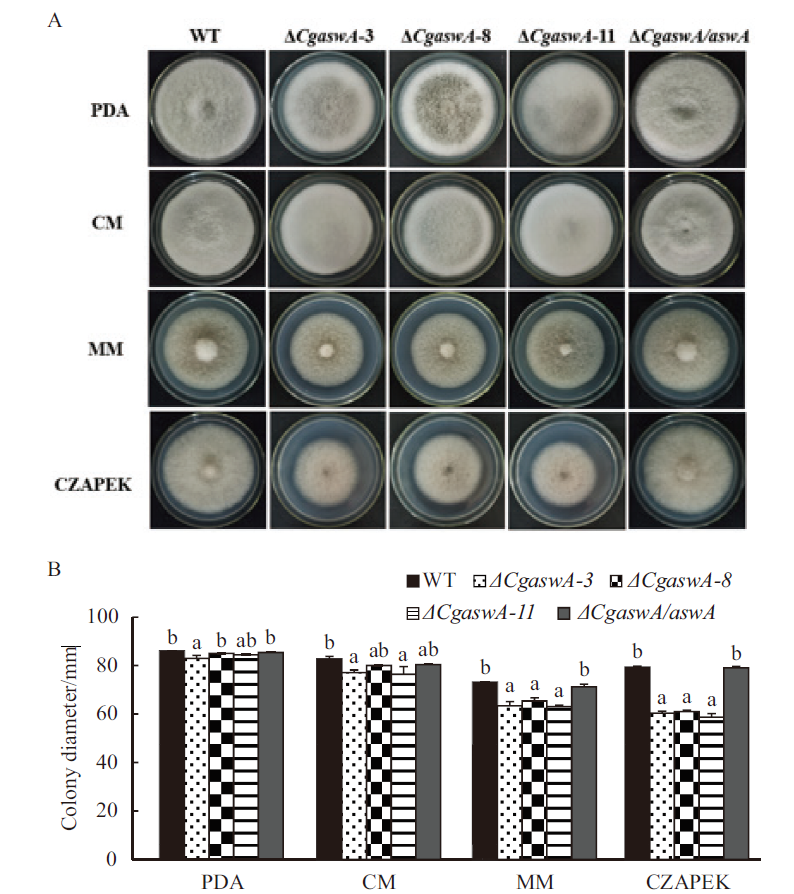
Fig. 4 Comparison of strains’ growth on four media A:Growth of strains on four media. B:Statistical analysis of colony diameter. a,b:Significant level,P<0.05
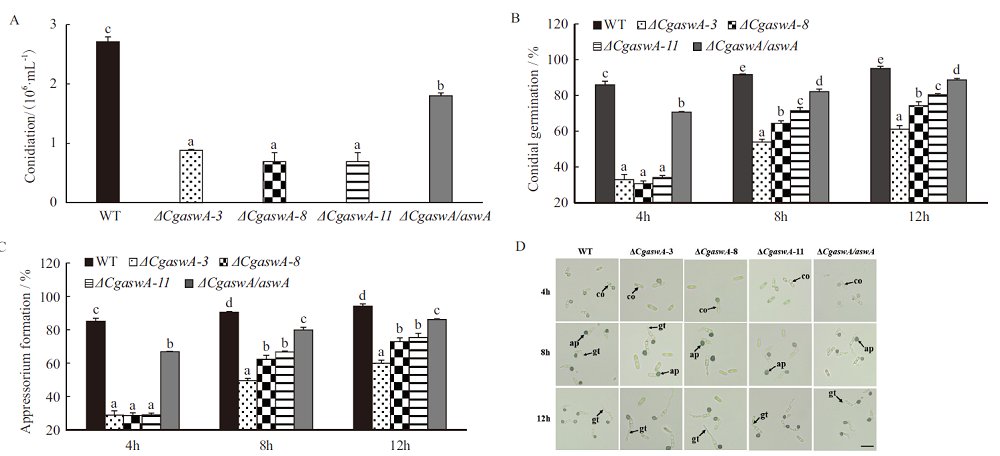
Fig. 5 Sporulation,germination and appressorium formation of wild type,ΔCgaswA and ΔCgaswA/aswA A:Statistical results of conidia yield. B:Statistical results of conidia germination rate. C:Statistical results of appressorium formation rate. D:Conidial germination and appressorium formation at 4 h,8 h and 12 h;co:conidium,gt:germ tube,ap:appressorium;a,b,c,d:significant level,P<0.05. Scale bars = 20 µm
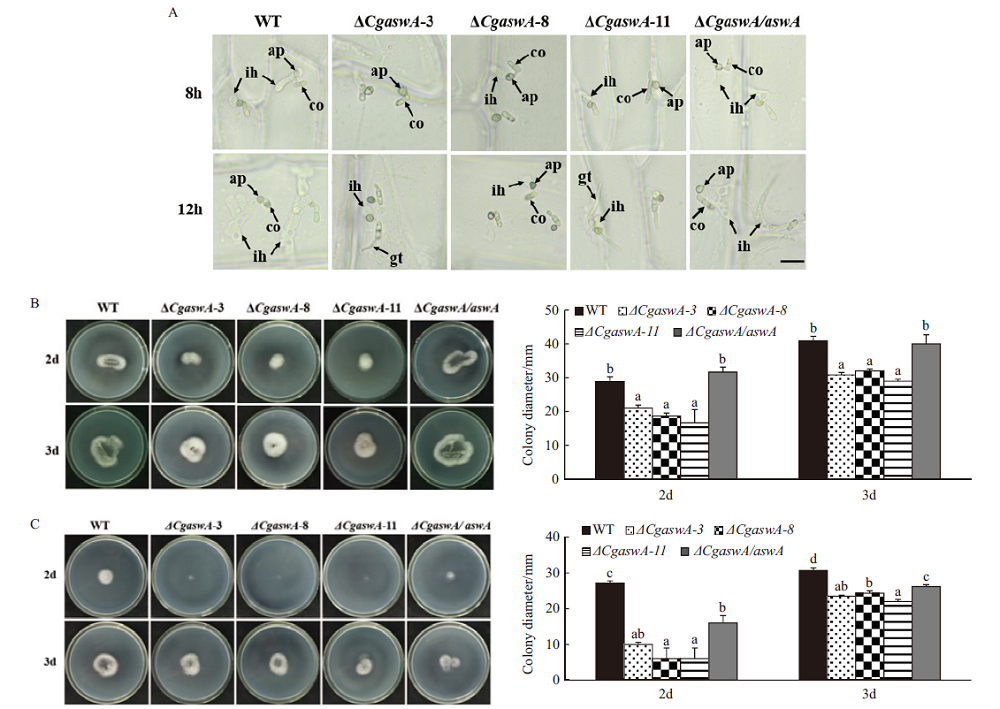
Fig. 6 Onion epidermis infection and cellophane penetration assays A:Appressorium infection assay of different strains on onion epidermis. B:Cellophane penetration assay of conidia suspensions. C:Cellophane penetration assay of mycelial plugs;co:conidium,ap:appressorium,ih:invasive hyphae;a,b,c,d:significant level,P<0.05;scale bars = 20 µm
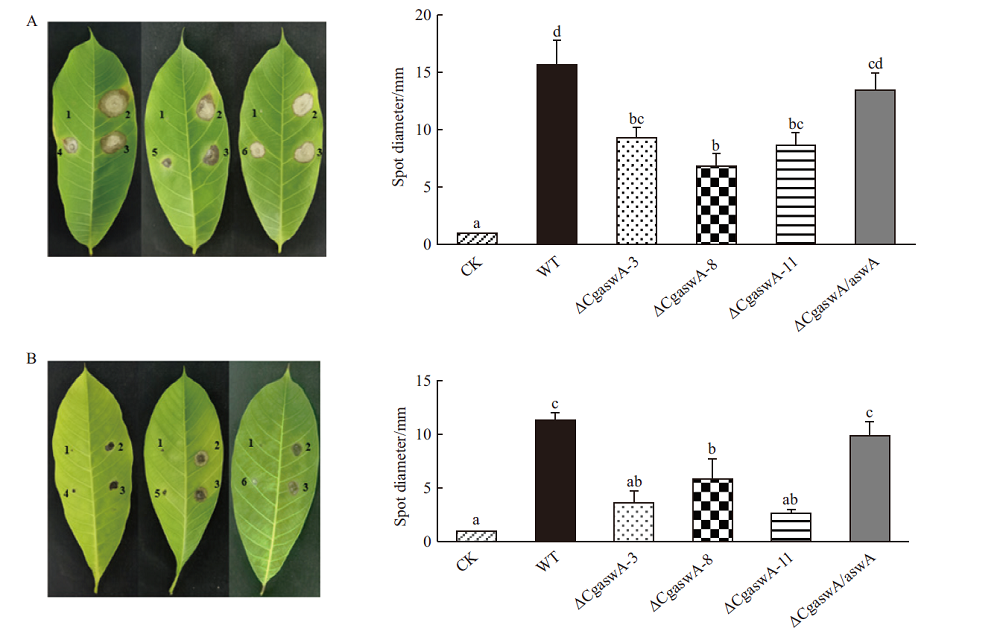
Fig. 7 Pathogenicity analyses of wild type,ΔCgaswA and ΔCgaswA/aswA A:Symptoms on rubber leaves at 4 d post-inoculation using mycelial plugs and statistical analyses of the diameters of disease spots. B:Symptomss on rubber leaves at 5 d post-inoculation using conidial suspensions and statistical analyses of the diameters of disease spots. 1:CK;2:WT;3:ΔCgaswA/ aswA;4:ΔCgaswA-3;5:ΔCgaswA-8;6:ΔCgaswA-11;a,b,c,d:significant level,P<0.05
| [1] | 韦周晓. 我国天然橡胶产业发展的困境及其对策分析[J]. 南方农业, 2020, 14(6):105-106. |
| Wei ZX. The predicament and countermeasure analysis of the development of natural rubber industry in China[J]. South China Agric, 2020, 14(6):105-106. | |
| [2] | 韩长志. 胶孢炭疽菌侵染过程相关基因研究进展[J]. 广东农业科学, 2014, 41(9):165-169. |
| Han CZ. Research advances on genes in infection process of Colletotrichum gloeosporioides[J]. Guangdong Agric Sci, 2014, 41(9):165-169. | |
| [3] |
Guyot J, Ntawanga Omanda E, Pinard F. Some epidemiological investigations on Colletotrichum leaf disease on rubber tree[J]. Crop Prot, 2005, 24(1):65-77.
doi: 10.1016/j.cropro.2004.06.009 URL |
| [4] | 韩长志. 胶孢炭疽病菌的研究进展[J]. 华北农学报, 2012, 27(S1):386-389. |
| Han CZ. Study on progress in Colletotrichum gloeosporioides[J]. Acta Agric Boreali Sin, 2012, 27(S1):386-389. | |
| [5] |
Turrà D, Segorbe D, Di Pietro A. Protein kinases in plant-pathogenic fungi:conserved regulators of infection[J]. Annu Rev Phytopathol, 2014, 52:267-288.
doi: 10.1146/phyto.2014.52.issue-1 URL |
| [6] | Latchman DS. Families of DNA binding transcription factors[M]// Eukaryotic transcription factors. Amsterdam:Elsevier, 2008:96-160. |
| [7] | 张莉林. 294个稻瘟病菌转录因子基因的敲除和功能分析[D]. 杭州:浙江大学, 2013. |
| Zhang LL. Knock-out and functional analysis of 294 transcription factor genes in Magnaporthe oryzae[D]. Hangzhou:Zhejiang University, 2013. | |
| [8] |
MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators:the zinc cluster proteins[J]. Microbiol Mol Biol Rev, 2006, 70(3):583-604.
doi: 10.1128/MMBR.00015-06 URL |
| [9] |
Lu J, Cao H, Zhang L, et al. Systematic analysis of Zn2Cys6 transcription factors required for development and pathogenicity by high-throughput gene knockout in the rice blast fungus[J]. PLoS Pathog, 2014, 10(10):e1004432.
doi: 10.1371/journal.ppat.1004432 URL |
| [10] |
Zhang L, Lubbers RJ, Simon A, et al. A novel Zn2 Cys6 transcription factor BcGaaR regulates D-galacturonic acid utilization in Botrytis cinerea[J]. Mol Microbiol, 2016, 100(2):247-262.
doi: 10.1111/mmi.2016.100.issue-2 URL |
| [11] | Felenbok B, Flipphi M, Nikolaev I. Ethanol catabolism in Aspergil-lus nidulans:a model system for studying gene regulation[J]. Prog Nucleic Acid Res Mol Biol, 2001, 69:149-204. |
| [12] |
Zhao CZ, Waalwijk C, de Wit PJ, et al. EBR1, a novel Zn(2)Cys(6)transcription factor, affects virulence and apical dominance of the hyphal tip in Fusarium graminearum[J]. Mol Plant Microbe Interact, 2011, 24(12):1407-1418.
doi: 10.1094/MPMI-06-11-0158 URL |
| [13] |
Boyce KJ, McLauchlan A, Schreider L, et al. Intracellular growth is dependent on tyrosine catabolism in the dimorphic fungal pathogen Penicillium marneffei[J]. PLoS Pathog, 2015, 11(3):e1004790.
doi: 10.1371/journal.ppat.1004790 URL |
| [14] |
Chang PK, Scharfenstein LL, Li RW, et al. Aspergillus flavus aswA, a gene homolog of Aspergillus nidulans oefC, regulates sclerotial development and biosynjournal of sclerotium-associated secondary metabolites[J]. Fungal Genet Biol, 2017, 104:29-37.
doi: 10.1016/j.fgb.2017.04.006 URL |
| [15] | 吴曼莉, 李晓宇, 张楠, 等. 胶孢炭疽菌CgRGS2基因的克隆及生物学功能[J]. 微生物学报, 2017, 57(1):66-76. |
| Wu ML, Li XY, Zhang N, et al. Gene cloning and biological function of CgRGS2 in Colletotrichum gloeosporioides[J]. Acta Microbiol Sin, 2017, 57(1):66-76. | |
| [16] |
Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea[J]. Plant Cell, 1993, 5(11):1575.
pmid: 8312740 |
| [17] |
Sweigard JA, Carroll AM, Kang S, et al. Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus[J]. Plant Cell, 1995, 7(8):1221-1233.
pmid: 7549480 |
| [18] | Lee B, Han S, Choi H, et al. Screening of growth- or development-related genes by using genomic library with inducible promoter in Aspergillus nidulans[J]. The Journal of Microbiology, 2005, 43(6):523-528. |
| [19] |
Son H, Seo YS, Min K, et al. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum[J]. PLoS Pathog, 2011, 7(10):e1002310.
doi: 10.1371/journal.ppat.1002310 URL |
| [20] |
Vienken K, Fischer R. The Zn(II)2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans[J]. Mol Microbiol, 2006, 61(2):544-554.
pmid: 16780567 |
| [21] |
Vienken K, Scherer M, Fischer R. The Zn(II)2Cys6 putative Aspergillus nidulans transcription factor repressor of sexual development inhibits sexual development under low-carbon conditions and in submersed culture[J]. Genetics, 2005, 169(2):619-630.
pmid: 15520269 |
| [22] | 张文琦. 大丽轮枝菌Fungaltrans转录因子VdFTF1调控致病性功能研究[D]. 北京:中国农业科学院, 2017. |
| Zhang WQ. Study on pathogenic mechanisms of fungaltrans transcription factor VdFTF1 in Verticillium dahliae[D]. Beijing:Chinese Academy of Agricultural Sciences, 2017. | |
| [23] | 曹慧娟. 稻瘟病菌Zn2Cys6和bHLH家族转录因子功能分析[D]. 杭州:浙江大学, 2015. |
| Cao HJ. Functional analyses of Magnaporthe orvzae Zn2Cys6 and bHLH transcription factors[D]. Hangzhou:Zhejiang University, 2015. | |
| [24] | 何锋. 梨树腐烂病菌的转录组分析及两个转录因子的功能研究[D]. 南京:南京农业大学, 2015. |
| He F. Transcriptome analysis of Valsa pyri and functional characterizations of two transcription factors[D]. Nanjing:Nanjing Agricultural University, 2015. | |
| [25] |
Sun Y, Wang Y, Tian C. bZIP transcription factor CgAP1 is essential for oxidative stress tolerance and full virulence of the poplar anthracnose fungus Colletotrichum gloeosporioides[J]. Fungal Genet Biol, 2016, 95:58-66.
doi: 10.1016/j.fgb.2016.08.006 URL |
| [26] |
Alkan N, Meng X, Friedlander G, et al. Global aspects of pacC regulation of pathogenicity genes in Colletotrichum gloeosporioides as revealed by transcriptome analysis[J]. Mol Plant Microbe Interact, 2013, 26(11):1345-1358.
doi: 10.1094/MPMI-03-13-0080-R URL |
| [27] |
Ment D, Alkan N, Luria N, et al. A role of AREB in the regulation of PACC-dependent acid-expressed-genes and pathogenicity of Colletotrichum gloeosporioides[J]. Mol Plant Microbe Interactions, 2015, 28(2):154-166.
doi: 10.1094/MPMI-09-14-0252-R URL |
| [28] |
Li XY, Ke ZJ, Yu XJ, et al. Transcription factor CgAzf1 regulates melanin production, conidial development and infection in Colletotrichum gloeosporioides[J]. Antonie Van Leeuwenhoek, 2019, 112(7):1095-1104.
doi: 10.1007/s10482-019-01243-1 URL |
| [29] |
Dubey AK, Barad S, Luria N, et al. Cation-stress-responsive transcription factors SltA and CrzA regulate morphogenetic processes and pathogenicity of Colletotrichum gloeosporioides[J]. PLoS One, 2016, 11(12):e0168561.
doi: 10.1371/journal.pone.0168561 URL |
| [1] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [2] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [3] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [4] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [5] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [6] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [7] | GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L. [J]. Biotechnology Bulletin, 2023, 39(6): 217-232. |
| [8] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [9] | PAN Guo-qiang, WU Si-yuan, LIU Lu, GUO Hui-ming, CHENG Hong-mei, SU Xiao-feng. Construction and Preliminary Analysis of Verticillim dahliae Mutant Library [J]. Biotechnology Bulletin, 2023, 39(5): 112-119. |
| [10] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [11] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [12] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| [13] | HU Ming-yue, YANG Yu, GUO Yang-dong, ZHANG Xi-chun. Functional Analysis of SlMYB96 Gene in Tomato Under Cold Stress [J]. Biotechnology Bulletin, 2023, 39(4): 236-245. |
| [14] | ZHANG Le-le, WANG Guan, LIU Feng, HU Han-qiao, REN Lei. Isolation, Identification and Biocontrol Mechanism of an Antagonistic Bacterium Against Anthracnose on Mango Caused by Colletotrichum gloeosporioides [J]. Biotechnology Bulletin, 2023, 39(4): 277-287. |
| [15] | GE Yan-rui, ZHAO Ran, XU Jing, LI Ruo-fan, HU Yun-tao, LI Rui-li. Advances in the Development and Regulation of Vascular Cambium [J]. Biotechnology Bulletin, 2023, 39(3): 13-25. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||