








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (9): 191-202.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1445
Previous Articles Next Articles
HONG Jun( ), WEI Xia-yi, JI Bing-jie, YE Yan-xin, CHENG Tian-ci
), WEI Xia-yi, JI Bing-jie, YE Yan-xin, CHENG Tian-ci
Received:2020-11-25
Online:2021-09-26
Published:2021-10-25
HONG Jun, WEI Xia-yi, JI Bing-jie, YE Yan-xin, CHENG Tian-ci. Change of Differentially Expressed Genes and SNP Before or After Pseudomonas aeruginosa Resistance to Tachyplesin I[J]. Biotechnology Bulletin, 2021, 37(9): 191-202.

Fig. 1 GO enrichment analysis of differentially expressed genes between PA1.2620 vs. PA-99 strains. The abscissa is -log10(P-value). The bigger the value of the abscissa,the more reliable the enrichment significance of differentially expressed genes in the functional annotation results. P-value<0.05 is the significant difference,and P-value<0.01 is the most significant difference. The ordinate is GO term. The same below
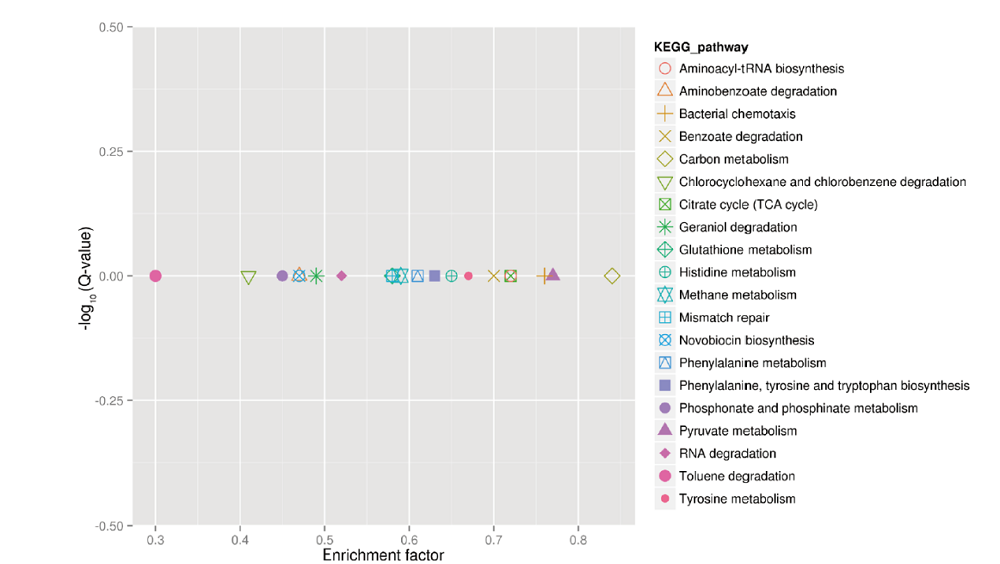
Fig.2 KEGG pathway enrichment analysis of differentially expressed genes between PA1.2620 vs. PA-99 strains Each graph in the figure represents one KEGG pathway,whose name is shown in the legend on the right. The abscissa is the enrichment factor,enrichment factor = amount of all genes/amount of DEGs enriched in the pathway in the background gene set. The smaller the enrichment factor,the more significant the enrichment level of differentially expressed genes in this pathway. The ordinate is -log10(Q-value),where Q-value is the P-value after the correction of multiple hypothesis testing. Therefore,the larger the ordinate,the more reliable the enrichment significance of differentially expressed genes in this pathway. It is generally believed that Q-value<0.05 is the significance difference,Q-value<0.01 is the most significant difference. The same below
| PA-99突变株样品 Sample of PA-99 mutant | Number of SNP | Transition | Transversion | Heterozygosity |
|---|---|---|---|---|
| T09 | 139 | 75.54% | 23.74% | 0.72% |
| T10 | 226 | 86.73% | 12.83% | 0.44% |
| T11 | 126 | 72.22% | 26.98% | 0.79% |
Table 1 Statistical table of SNP sites
| PA-99突变株样品 Sample of PA-99 mutant | Number of SNP | Transition | Transversion | Heterozygosity |
|---|---|---|---|---|
| T09 | 139 | 75.54% | 23.74% | 0.72% |
| T10 | 226 | 86.73% | 12.83% | 0.44% |
| T11 | 126 | 72.22% | 26.98% | 0.79% |
| #ID or Gene name | Protein name | log2FC | Regulated | Start | End | SNP number | SNP |
|---|---|---|---|---|---|---|---|
| pvdL | Peptide synthase | 1.15621 | Up | 2707666 | 2720694 | 2 | 2712019C>A;2715424T>C |
| Novel_244 | — | -1.71631 | Down | 1720641 | 1732256 | 1 | 1732254T>C |
| PA1938 | Hypothetical protein | -1.95428 | Down | 2118926 | 2119747 | 1 | 2119153T>C |
| Novel_497 | — | 1.95302 | Up | 3672989 | 3674050 | 1 | 3673743G>A |
| Novel_745 | — | 2.58303 | Up | 5130955 | 5135726 | 1 | 5130955T>C |
| Novel_797 | — | -1.77794 | Down | 5343486 | 5344904 | 1 | 5344709G>A |
| Novel_840 | — | -2.52117 | Down | 5621609 | 5624734 | 1 | 5621617G>A |
| PA1428a | Hypothetical protein | 1.07562 | Up | 1552566 | 1554155 | 1 | 1553722G>A |
| PA0193 | Hypothetical protein | 1.186966 | Up | 221585 | 222487 | 1 | 222358C>T |
| exoY | Adenylate cyclase | -1.75024 | Down | 2410344 | 2411480 | 1 | 2411150G>T |
| PA2451 | Hypothetical protein | 1.56984 | Up | 2752866 | 2754445 | 1 | 2754392G>A |
| cspD | Cold-shock protein CspD | -1.0514 | Down | 2965201 | 2965473 | 1 | 2965461C>T |
| PA2712 | Hypothetical protein | -1.01673 | Down | 3066296 | 3067159 | 1 | 3066971A>G |
| PA3380 | Hypothetical protein | 2.27038 | Up | 3787021 | 3787479 | 1 | 3787410G>A |
| opr86 | Outer membrane protein Opr86 | -1.51418 | Down | 4085062 | 4087455 | 1 | 4085961C>G |
| cupB3 | Usher CupB3 | -1.59417 | Down | 4565421 | 4567955 | 1 | 4565913A>G |
| fepD | Ferric enterobactin transporter FepD | 1.26932 | Up | 4655368 | 4656390 | 1 | 4656097T>C |
| pagL | Lipid A 3-O-deacylase | -2.39699 | Down | 5229459 | 5229980 | 1 | 5229837C>T |
| pmrB | Two-component regulator system Signal sensor kinase PmrB | 1.00016 | Up | 5364760 | 5366193 | 1 | 5364817G>T |
| PA4837 | Hypothetical protein | 1.411356 | Up | 5427716 | 5429842 | 1 | 5428274G>A |
| bioD | ATP-dependent dethiobiotin Synthetase BioD | 1.47708 | Up | 563549 | 564235 | 1 | 563798C>T |
| vreR | Sigma factor regulator VreR | 1.68468 | Up | 735487 | 736446 | 1 | 735564C>T |
Table 2 SNP analysis of differentially expressed genes in PA1.2620 vs. PA-99 strains
| #ID or Gene name | Protein name | log2FC | Regulated | Start | End | SNP number | SNP |
|---|---|---|---|---|---|---|---|
| pvdL | Peptide synthase | 1.15621 | Up | 2707666 | 2720694 | 2 | 2712019C>A;2715424T>C |
| Novel_244 | — | -1.71631 | Down | 1720641 | 1732256 | 1 | 1732254T>C |
| PA1938 | Hypothetical protein | -1.95428 | Down | 2118926 | 2119747 | 1 | 2119153T>C |
| Novel_497 | — | 1.95302 | Up | 3672989 | 3674050 | 1 | 3673743G>A |
| Novel_745 | — | 2.58303 | Up | 5130955 | 5135726 | 1 | 5130955T>C |
| Novel_797 | — | -1.77794 | Down | 5343486 | 5344904 | 1 | 5344709G>A |
| Novel_840 | — | -2.52117 | Down | 5621609 | 5624734 | 1 | 5621617G>A |
| PA1428a | Hypothetical protein | 1.07562 | Up | 1552566 | 1554155 | 1 | 1553722G>A |
| PA0193 | Hypothetical protein | 1.186966 | Up | 221585 | 222487 | 1 | 222358C>T |
| exoY | Adenylate cyclase | -1.75024 | Down | 2410344 | 2411480 | 1 | 2411150G>T |
| PA2451 | Hypothetical protein | 1.56984 | Up | 2752866 | 2754445 | 1 | 2754392G>A |
| cspD | Cold-shock protein CspD | -1.0514 | Down | 2965201 | 2965473 | 1 | 2965461C>T |
| PA2712 | Hypothetical protein | -1.01673 | Down | 3066296 | 3067159 | 1 | 3066971A>G |
| PA3380 | Hypothetical protein | 2.27038 | Up | 3787021 | 3787479 | 1 | 3787410G>A |
| opr86 | Outer membrane protein Opr86 | -1.51418 | Down | 4085062 | 4087455 | 1 | 4085961C>G |
| cupB3 | Usher CupB3 | -1.59417 | Down | 4565421 | 4567955 | 1 | 4565913A>G |
| fepD | Ferric enterobactin transporter FepD | 1.26932 | Up | 4655368 | 4656390 | 1 | 4656097T>C |
| pagL | Lipid A 3-O-deacylase | -2.39699 | Down | 5229459 | 5229980 | 1 | 5229837C>T |
| pmrB | Two-component regulator system Signal sensor kinase PmrB | 1.00016 | Up | 5364760 | 5366193 | 1 | 5364817G>T |
| PA4837 | Hypothetical protein | 1.411356 | Up | 5427716 | 5429842 | 1 | 5428274G>A |
| bioD | ATP-dependent dethiobiotin Synthetase BioD | 1.47708 | Up | 563549 | 564235 | 1 | 563798C>T |
| vreR | Sigma factor regulator VreR | 1.68468 | Up | 735487 | 736446 | 1 | 735564C>T |
| sRNA ID | Gene length/nt | log2FC | Regulated | sRNA ID | Gene length/nt | log2FC | Regulated |
|---|---|---|---|---|---|---|---|
| Novel_33 | 188 | -3.101050802 | Down | Novel_346 | 136 | -1.138061426 | Down |
| Novel_377 | 378 | -2.065876992 | Down | Novel_593 | 127 | -1.434477351 | Down |
| Novel_583 | 78 | -3.461144761 | Down | Novel_683 | 153 | -1.371013476 | Down |
| Novel_584 | 116 | -2.305895532 | Down | Novel_344 | 60 | 1.764469442 | Up |
| Novel_6 | 94 | -1.576755833 | Down | Novel_5 | 284 | -1.660811503 | Down |
| Novel_853 | 117 | -1.507421364 | Down |
Table 3 sRNA of differentially expressed genes in PA1.2620 vs. PA-99 strains
| sRNA ID | Gene length/nt | log2FC | Regulated | sRNA ID | Gene length/nt | log2FC | Regulated |
|---|---|---|---|---|---|---|---|
| Novel_33 | 188 | -3.101050802 | Down | Novel_346 | 136 | -1.138061426 | Down |
| Novel_377 | 378 | -2.065876992 | Down | Novel_593 | 127 | -1.434477351 | Down |
| Novel_583 | 78 | -3.461144761 | Down | Novel_683 | 153 | -1.371013476 | Down |
| Novel_584 | 116 | -2.305895532 | Down | Novel_344 | 60 | 1.764469442 | Up |
| Novel_6 | 94 | -1.576755833 | Down | Novel_5 | 284 | -1.660811503 | Down |
| Novel_853 | 117 | -1.507421364 | Down |
| Gene ID | Protein name | log2FC | Regulated | Start | End | SNP number | SNP |
|---|---|---|---|---|---|---|---|
| pvdL | Peptide synthase | 1.156212 | Up | 2707666 | 2720694 | 2 | 2712019C>A;2715424T>C |
| vreR | Sigma factor regulator VreR | 1.684682 | Up | 735487 | 736446 | 1 | 735564C>T |
| Novel_497 | — | 1.953026 | Up | 3672989 | 3674050 | 1 | 3673743G>A |
| Novel_745 | — | 2.583033 | Up | 5130955 | 5135726 | 1 | 5130955T>C |
| opr86 | Outer membrane protein Opr86 | -1.51418 | Down | 4085062 | 4087455 | 1 | 4085961C>G |
| cupB3 | Usher CupB3 | -1.59417 | Down | 4565421 | 4567955 | 1 | 4565913A>G |
| pmrB | Two-component regulator System signal sensor kinase | 1.000160 | Up | 5364760 | 5366193 | 1 | 5364817G>T |
| Novel_840 | — | -2.52116 | Down | 5621609 | 5624734 | 1 | 5621617G>A |
| PA2451 | Hypothetical protein | 1.569849 | Up | 2752866 | 2754445 | 1 | 2754392G>A |
| PA2712 | Hypothetical protein | -1.01672 | Down | 3066296 | 3067159 | 1 | 3066971A>G |
Table 4 SNP analysis of sRNA target genes
| Gene ID | Protein name | log2FC | Regulated | Start | End | SNP number | SNP |
|---|---|---|---|---|---|---|---|
| pvdL | Peptide synthase | 1.156212 | Up | 2707666 | 2720694 | 2 | 2712019C>A;2715424T>C |
| vreR | Sigma factor regulator VreR | 1.684682 | Up | 735487 | 736446 | 1 | 735564C>T |
| Novel_497 | — | 1.953026 | Up | 3672989 | 3674050 | 1 | 3673743G>A |
| Novel_745 | — | 2.583033 | Up | 5130955 | 5135726 | 1 | 5130955T>C |
| opr86 | Outer membrane protein Opr86 | -1.51418 | Down | 4085062 | 4087455 | 1 | 4085961C>G |
| cupB3 | Usher CupB3 | -1.59417 | Down | 4565421 | 4567955 | 1 | 4565913A>G |
| pmrB | Two-component regulator System signal sensor kinase | 1.000160 | Up | 5364760 | 5366193 | 1 | 5364817G>T |
| Novel_840 | — | -2.52116 | Down | 5621609 | 5624734 | 1 | 5621617G>A |
| PA2451 | Hypothetical protein | 1.569849 | Up | 2752866 | 2754445 | 1 | 2754392G>A |
| PA2712 | Hypothetical protein | -1.01672 | Down | 3066296 | 3067159 | 1 | 3066971A>G |
| [1] |
Nakamura T, Furunaka H, Miyata T, et al. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab(Tachypleus tridentatus)isolation and chemical structure[J]. Journal of Biological Chemistry, 1988, 263:16709-16713.
pmid: 3141410 |
| [2] |
Anaya-López1 JL, López-Meza JE. Bacterial resistance to cationic antimicrobial peptides[J]. Critical Reviews in Microbiology, 2013, 39(2):180-195.
doi: 10.3109/1040841X.2012.699025 pmid: 22799636 |
| [3] | Cai Y, Chai D, Wang R, et al. Colistin resistance of Acinetobacter baumannii:clinical reports, mechanisms and antimicrobial strategies[J]. Antimicrobial Agents and Chemotherapy, 2012, 67:1607-1615. |
| [4] |
Hong J, Hu JY, Ke F. Experimental induction of bacterial resistance to the antimicrobial peptide tachyplesin I and investigation of the resistance mechanisms[J]. Antimicrobial Agents and Chemotherapy, 2016, 60(10):6067-6075.
doi: 10.1128/AAC.00640-16 pmid: 27480861 |
| [5] | 洪军, 胡建业, 刘坤, 等. 绿脓杆菌对鲎素耐受性特点及耐受性机制[J]. 微生物学报, 2018, 58(9):1593-1604. |
| Hong J, Hu JY, Liu K, et al. Characteristics and resistance of tachyplesin-I resistance in Pseudomonas aeruginosa[J]. Acta Microbiologica Sinica, 2018, 58(9):1593-1604. | |
| [6] | Peng T, Kan J, Lun JS, et al. Identification of novel sRNAs involved in oxidative stress response in the fish pathogen Vibrio alginolyticus by transcriptome analysis[J]. Journal of Fish Diseases, 2019, 42(2):1-15. |
| [7] |
Barnhill EC, Crucello A, Houserova D, et al. Characterization of novel small RNAs(sRNAs)contributing to the desiccation response of Salmonella enterica serovar Typhimurium[J]. RNA Biology, 2019, 16(11):1643-1657.
doi: 10.1080/15476286.2019.1653680 URL |
| [8] | 叶中杨, 邱怀雨, 祝丙华, 等. sRNA调控细菌耐药相关基因表达研究进展[J]. 中国生物工程杂志, 2018, 38(7):89-93. |
| Ye ZY, Qiu HY, Zhu BH, et al. Research progress of sRNA regulates the expression of genes in related with bacterial resistance[J]. China Biotechnology, 2018, 38(7):89-93. | |
| [9] |
Miller CL, Manuel R, Rajasekhar KSL, et al. RsmW, Pseudomonas aeruginosa small non-coding RsmA-binding RNA upregulated in biofilm versus planktonic growth conditions[J]. BMC Microbiology, 2016, 16(1):155.
doi: 10.1186/s12866-016-0771-y URL |
| [10] | 刘翠翠. 福氏志贺菌耐药株的转录组测定及sRNA差异分析[D]. 北京:中国农业科学院, 2014. |
| Liu CC. The transcriptome sequencing analysis and sRNA difference analysis in drug-resistant Shigella flexneri[D]. Beijing:Chinese Academy of Agricultural Sciences, 2014. | |
| [11] |
Zhang YF, Han K, Chandler CE, et al. Probing the sRNA regulatory landscapeof P. aeruginosa:post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility[J]. Molecular Microbiology, 2017, 106(6):919-937.
doi: 10.1111/mmi.2017.106.issue-6 URL |
| [12] | 张栋, 高风英, 卢迈新, 等. 尼罗罗非鱼β2 m基因SNP位点和单倍型与无乳链球菌抗性的关联分析[J]. 水生生物学报, 2018, 42(5):903-912. |
| Zhang D, Gao FY, Lu MX, et al. Association analysis of SNP locus and haplotype of β2m gene in Nile tilapia(Oreochromis niloticus)with its resistance to Streptococcus agalactiae[J]. Acta Hydrobiologica Sinica, 2018, 42(5):903-912. | |
| [13] |
Wang L, Feng Z, Wang X, et al. DEGseq:an R package for identifying differentially expressed genes from RNAseq data[J]. Bioinformatics, 2010, 26:136-138.
doi: 10.1093/bioinformatics/btp612 URL |
| [14] | Ashburner M, Ball CA, Blake JA, et al. Gene ontology:tool for the unification of biology[J]. Nature Genetics, 2000, 25(1):2529. |
| [15] | Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome[J]. Nucleic Acids Research, 2004, 32:D277280. |
| [16] |
Winsor GL, Griffiths EJ, Lo R, et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database[J]. Nucleic Acids Research, 2016, 44:D646-D653.
doi: 10.1093/nar/gkv1227 URL |
| [17] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method[J]. Methods, 2001, 25:402-408.
pmid: 11846609 |
| [18] |
McPhee JB, Bains M, Winsor G, et al. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa[J]. Journal of Bacteriology, 2006, 188:3995-4006.
doi: 10.1128/JB.00053-06 URL |
| [19] |
Moskowitz SM, Brannon MK, Dasgupta N, et al. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients[J]. Antimicrobial Agents and Chemotherapy, 2012, 56(2):1019-1030.
doi: 10.1128/AAC.05829-11 pmid: 22106224 |
| [20] |
Shepherd J, Ibba M. Direction of aminoacylated transfer RNAs into antibiotic synjournal and peptidoglycan-mediated antibiotic resistance[J]. FEBS Letters, 2013, 587(18):2895-2904.
doi: 10.1016/j.febslet.2013.07.036 URL |
| [21] |
Ernst CM, Peschel A. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids[J]. Molecular Microbiology, 2011, 80:290-299.
doi: 10.1111/mmi.2011.80.issue-2 URL |
| [22] | 许达, 宗树成, 胡森, 等. 细菌sRNA代谢调控的研究进展[J]. 中国预防兽医学报, 2017, 39(1):80-84. |
| Xu D, Zong SC, Hu S, et al. Research progress on regulation of Bacterial sRNA metabolism[J]. Chinese Journal of Preventive Veterinary Medicine, 2017, 39(1):80-84. |
| [1] | WU Hao, LIU Zi-wei, ZHENG Ying, DAI Ya-wen, SHI Quan. Study on the Heterogeneity of Human Gingival Mesenchymal Stem Cells at Single Cell Level [J]. Biotechnology Bulletin, 2023, 39(7): 325-332. |
| [2] | CUI Xue-qiang, HUANG Chang-yan, DENG Jie-ling, LI Xian-min, LI Xiu-ling, ZHANG Zi-bin. SNP Markers Development and Genetic Relationship Analysis of Dendrobium Germplasms Using SLAF-seq Technology [J]. Biotechnology Bulletin, 2023, 39(6): 141-148. |
| [3] | ZHAO Jin-ling, AN Lei, REN Xiao-liang. Development of Single Cell Transcriptome Sequencing Technology and Its Application in Caenorhabditis elegans [J]. Biotechnology Bulletin, 2023, 39(6): 158-170. |
| [4] | XIAO Xiao-jun, CHEN Ming, HAN De-peng, YU Pao-lan, ZHENG Wei, XIAO Guo-bin, ZHOU Qing-hong, ZHOU Hui-wen. Genome Wide Association Analysis of Thousand Seed Weight in Brassica napus L. [J]. Biotechnology Bulletin, 2023, 39(3): 143-151. |
| [5] | HE Li-na, FENG Yuan, SHI Hui-min, YE Jian-ren. Screening and Identification of Endophytic Bacteria with Nematicidal Activity Against Bursaphelenchus xylophilus in Pinus massoniana [J]. Biotechnology Bulletin, 2022, 38(8): 159-166. |
| [6] | ZHOU Xiao-nan, XU Jin-qing, LEI Yu-qing, WANG Hai-qing. Development of SNP Markers in Medicago archiducis-nicolai Based on GBS-seq [J]. Biotechnology Bulletin, 2022, 38(4): 303-310. |
| [7] | HU Shan, LIANG Wei-qu, HUANG Hao, XU Cong, LUO Hua-jian, HU Chu-wei, HUANG Xiao-yan, CHEN Shi-li. Screening,Identification and Antagonism of Phosphate-Solubilizing Bacteria from the Compost Chinese Medicinal Herbal Residues [J]. Biotechnology Bulletin, 2022, 38(3): 92-102. |
| [8] | ZHENG Qing-bo, YE Na, ZHANG Xiao-lan, BAO Peng-jia, WANG Fu-bin, REN Wen-wen, LIAO Yue-jiao, YAN Ping, PAN He-ping. Identification of Hair Follicle Cell Subsets and Bioinformatics Analysis of Characteristic Genes in Tianzhu White Yak During Catagen [J]. Biotechnology Bulletin, 2022, 38(10): 262-272. |
| [9] | LI Ling, YANG Li-xia, GUO Mei. Function of Transcription Factor CNR in the Ripening Process of Tomato Fruit [J]. Biotechnology Bulletin, 2021, 37(2): 51-62. |
| [10] | ZHANG Ting-huan, LONG Xi, GUO Zong-yi, CHAI Jie. miR-378 Promoting Lipogenesis and Identification of Target Genes [J]. Biotechnology Bulletin, 2021, 37(2): 80-87. |
| [11] | HE Jin-hua, MA Xiang, TANG Yan-qiong, WANG Dan, LI Hong, LIU Zhu. Identification and Function Study of a New Type of sRNA N155 from Aeromonas veronii [J]. Biotechnology Bulletin, 2021, 37(11): 267-275. |
| [12] | DENG Pu-rong, LIU Yong-bo. Review on the Synergistic Insect-resistant Application of RNAi and Bt-transgenic Technologies [J]. Biotechnology Bulletin, 2021, 37(10): 216-224. |
| [13] | YE Na, ZHANG Xiao-lan, BAO Peng-jia, WANG Xing-dong, YAN Ping, PAN He-ping. Single Cell Sequencing Technology and Its Application in Hair Follicle Development [J]. Biotechnology Bulletin, 2021, 37(10): 245-256. |
| [14] | ZHANG Ting-ting, TENG Li, CAI Xiao-yao, LONG Hui, LI Sheng-yu, LIU Hong-mei. Genomic Characterization of a Totiviridae from the Ustilaginoidea virens Strain GZ-14-07 [J]. Biotechnology Bulletin, 2020, 36(10): 80-87. |
| [15] | ZOU Kun, CUI Hong-yan, XUE Yuan, ZHANG Shao-wei, LU Li-li, ZHAO Zhi-hui, SU Ying. Correlation Between FSHR and ESR1 and Breeding Traits in Leizhou Black Duck [J]. Biotechnology Bulletin, 2019, 35(8): 118-126. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||