








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (10): 90-96.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1477
Previous Articles Next Articles
CHEN Li1( ), LU Xi1, YANG Hong-lan1, ZHANG Peng1,2(
), LU Xi1, YANG Hong-lan1, ZHANG Peng1,2( ), HE Zhi-xu1,2,3(
), HE Zhi-xu1,2,3( )
)
Received:2021-11-29
Online:2022-10-26
Published:2022-11-11
Contact:
ZHANG Peng,HE Zhi-xu
E-mail:2696711908@qq.com;peng12zhang@gmail.com;hzx@gmc.edu.cn
CHEN Li, LU Xi, YANG Hong-lan, ZHANG Peng, HE Zhi-xu. Methodological Research on Rapid Detection of CRISPR/Cas9-Mediated Gene Mutations in Cells Based on High-resolution Melting Technique[J]. Biotechnology Bulletin, 2022, 38(10): 90-96.

Fig.1 Analysis of high-resolution melting of wild-type mESCs,homozygous mutation mESCs and heterozygous mutation mESCs when the PCR product of wild-type mESCs is 394 bp

Fig.2 Analysis of high-resolution melting of wild-type mE-SCs,homozygous mutation mESCs and heterozygous mutation mESCs when the PCR product of wild-type mESCs is 91 bp
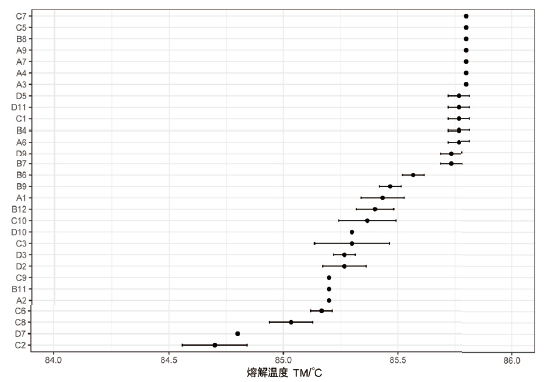
Fig.5 Results of melting temperature of 30 monoclones of mESCs edited by CRISPR/Cas9 techniqe The abscissa refers to the melting temperature,and the vertical axis refers to 30 different monoclones of mESCs,among which monoclones C7-B7 are wild-type mESCs,monoclones B6-C8 are heterozygous mutation mESCs,and monoclones D7 and C2 are homozygous mutantion mESCs
| 方法 Method | 检测SNP Detecting SNP | 灵敏度 Sensitivity | 检测周期 Detection period | 检测成本 Detection cost | 检测步骤 Detection steps | 高通量方法 High-throughput method |
|---|---|---|---|---|---|---|
| 酶错配切割方法 Mismatch cleavage assays | 能Yes | 低Low | 较长 A little long | 低Low | 繁琐Complex | 否No |
| DNA测序法Sanger sequencing | 能Yes | 高High | 长Long | 高High | 繁琐Complex | 否No |
| 高分辨率熔解曲线法HRMA | 能Yes | 高High | 短Short | 低Low | 简单Simple | 是Yes |
Table 1 Comparison of three methods of detecting gene products edited by CRISPR/Cas9 technique
| 方法 Method | 检测SNP Detecting SNP | 灵敏度 Sensitivity | 检测周期 Detection period | 检测成本 Detection cost | 检测步骤 Detection steps | 高通量方法 High-throughput method |
|---|---|---|---|---|---|---|
| 酶错配切割方法 Mismatch cleavage assays | 能Yes | 低Low | 较长 A little long | 低Low | 繁琐Complex | 否No |
| DNA测序法Sanger sequencing | 能Yes | 高High | 长Long | 高High | 繁琐Complex | 否No |
| 高分辨率熔解曲线法HRMA | 能Yes | 高High | 短Short | 低Low | 简单Simple | 是Yes |
| [1] |
Gasanov EV, Jędrychowska J, Pastor M, et al. An improved method for precise genome editing in zebrafish using CRISPR-Cas9 technique[J]. Mol Biol Rep, 2021, 48(2):1951-1957.
doi: 10.1007/s11033-020-06125-8 pmid: 33481178 |
| [2] |
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering[J]. Cell, 2014, 157(6):1262-1278.
doi: S0092-8674(14)00604-7 pmid: 24906146 |
| [3] | 胡廷栋, 王蒙, 刘晓蕊, 等. 利用CRISPR/Cas9建立PIN1基因敲除的成体神经干细胞系[J]. 南京医科大学学报:自然科学版, 2021, 41(4):483-488, 521. |
| Hu TD, Wang M, Liu XR, et al. Establishment of neural stem cells line with PIN1 gene knockout by CRISPR/Cas9[J]. J Nanjing Med Univ Nat Sci, 2021, 41(4):483-488, 521. | |
| [4] |
Druml B, Cichna-Markl M. High resolution melting(HRM)analysis of DNA—its role and potential in food analysis[J]. Food Chem, 2014, 158:245-254.
doi: 10.1016/j.foodchem.2014.02.111 URL |
| [5] | 温鹏强, 王国兵, 陈占玲, 等. 应用高分辨率熔解曲线分析快速筛查和诊断citrin缺陷导致的新生儿肝内胆汁淤积症[J]. 中华医学遗传学杂志, 2012, 29(2):167-171. |
| Wen PQ, Wang GB, Chen ZL, et al. Utilization of high-resolution melting analysis to screen patients with neonatal intrahepatic cholestasis caused by citrin deficiency[J]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi, 2012, 29(2):167-171. | |
| [6] |
Tricarico R, Crucianelli F, Alvau A, et al. High resolution melting analysis for a rapid identification of heterozygous and homozygous sequence changes in the MUTYH gene[J]. BMC Cancer, 2011, 11:305.
doi: 10.1186/1471-2407-11-305 pmid: 21777424 |
| [7] |
Marotta RV, Turri O, Morandi A, et al. High resolution melting analysis to genotype the most common variants in the HFE gene[J]. Clin Chem Lab Med, 2011, 49(9):1453-1457.
doi: 10.1515/CCLM.2011.237 pmid: 21627541 |
| [8] |
Bodnar GC, Martins HM, de Oliveira CF, et al. Comparison of HRM analysis and three REP-PCR genomic fingerprint methods for rapid typing of MRSA at a Brazilian hospital[J]. J Infect Dev Ctries, 2016, 10(12):1306-1317.
doi: 10.3855/jidc.7887 URL |
| [9] |
Chambliss AB, Resnick M, Petrides AK, et al. Rapid screening for targeted genetic variants via high-resolution melting curve analysis[J]. Clin Chem Lab Med, 2017, 55(4):507-516.
doi: 10.1515/cclm-2016-0603 pmid: 27732553 |
| [10] |
Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis[J]. Clin Chem, 2004, 50(10):1748-1754.
pmid: 15308590 |
| [11] |
Switzeny OJ, Christmann M, Renovanz M, et al. MGMT promoter methylation determined by HRM in comparison to MSP and pyrosequencing for predicting high-grade glioma response[J]. Clin Epigenetics, 2016, 8:49.
doi: 10.1186/s13148-016-0204-7 pmid: 27158275 |
| [12] |
Wang XM, Shi H, Zhou JJ, et al. Generation of rat blood vasculature and hematopoietic cells in rat-mouse chimeras by blastocyst complementation[J]. J Genet Genomics, 2020, 47(5):249-261.
doi: S1673-8527(20)30086-2 pmid: 32703661 |
| [13] | 张鹏, 杨红兰, 刘含, 等. 甲基结合蛋白1对小鼠胚胎干细胞增殖及克隆形态的影响[J]. 贵州医科大学学报, 2019, 44(6):621-625. |
| Zhang P, Yang HL, Liu H, et al. Effect of methyl-CpG binding domain protein 1 on the colony morphology and proliferation of mouse embryonic stem cells[J]. J Guizhou Med Univ, 2019, 44(6):621-625. | |
| [14] |
Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and Archaea[J]. Nature, 2012, 482(7385):331-338.
doi: 10.1038/nature10886 URL |
| [15] |
Nelles DA, Fang MY, O’Connell MR, et al. Programmable RNA tracking in live cells with CRISPR/Cas9[J]. Cell, 2016, 165(2):488-496.
doi: 10.1016/j.cell.2016.02.054 pmid: 26997482 |
| [16] | 龚美玲, 张琳琳, 郑翠侠. 利用CRISPR/Cas9系统对人A549肺癌细胞NRF2基因的稳定敲除及其功能研究[J]. 中国癌症杂志, 2019, 29(11):855-861. |
| Gong ML, Zhang LL, Zheng CX. Stable knockout of NRF2 gene in human A549 lung cancer cells by CRISPR/Cas9 system and its functional research[J]. China Oncol, 2019, 29(11):855-861. | |
| [17] |
Gupta D, Bhattacharjee O, Mandal D, et al. CRISPR-Cas9 system:a new-fangled dawn in gene editing[J]. Life Sci, 2019, 232:116636.
doi: 10.1016/j.lfs.2019.116636 URL |
| [18] |
Memi FN, Ntokou A, Papangeli I. CRISPR/Cas9 gene-editing:research technologies, clinical applications and ethical considerations[J]. Semin Perinatol, 2018, 42(8):487-500.
doi: 10.1053/j.semperi.2018.09.003 URL |
| [19] |
Janik E, Niemcewicz M, Ceremuga M, et al. Various aspects of a gene editing system-CRISPR-Cas9[J]. Int J Mol Sci, 2020, 21(24):9604.
doi: 10.3390/ijms21249604 URL |
| [20] | Vouillot L, Thélie A, Pollet N. Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases[J]. G3(Bethesda), 2015, 5(3):407-415. |
| [21] |
Zischewski J, Fischer R, Bortesi L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases[J]. Biotechnol Adv, 2017, 35(1):95-104.
doi: S0734-9750(16)30158-6 pmid: 28011075 |
| [22] |
Martín-Núñez GM, Gómez-Zumaquero JM, Soriguer F, et al. High resolution melting curve analysis of DNA samples isolated by different DNA extraction methods[J]. Clin Chimica Acta, 2012, 413(1/2):331-333.
doi: 10.1016/j.cca.2011.09.014 URL |
| [23] |
Pham QT, Raad S, Mangahas CL, et al. High-throughput assessment of mutations generated by genome editing in induced pluripotent stem cells by high-resolution melting analysis[J]. Cytotherapy, 2020, 22(10):536-542.
doi: S1465-3249(20)30754-4 pmid: 32768274 |
| [1] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [2] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [3] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [4] | ZHANG Xue-ping, LU Yu-qing, ZHANG Yue-qian, LI Xiao-juan. Advances in Plant Extracellular Vesicles and Analysis Techniques [J]. Biotechnology Bulletin, 2023, 39(5): 32-43. |
| [5] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [6] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [7] | ZHOU Xi-wen, CHENG Ke, ZHU Hong-liang. Research Progress in the Approaches to in vivo RNA Secondary Structure Profiling in Plants [J]. Biotechnology Bulletin, 2023, 39(2): 51-62. |
| [8] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [9] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [10] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [11] | GUO Wen-bo, LU Yang, SUI Li, ZHAO Yu, ZOU Xiao-wei, ZHANG Zheng-kun, LI Qi-yun. Preparation and Application of Polyclonal Antibodies Against Beauveria bassiana Mycovirus BbPmV-4 Coat Protein [J]. Biotechnology Bulletin, 2023, 39(10): 58-67. |
| [12] | LI Hui-jie, DONG Lian-hua, CHEN Gui-fang, LIU Si-yuan, YANG Jia-yi, YANG Jing-ya. Establishment of Droplet Digital PCR Assay for Quantitative Detection of Pseudomonas cocovenenans in Foods [J]. Biotechnology Bulletin, 2023, 39(1): 127-136. |
| [13] | CHEN Xiao-lin, LIU Yang-er, XU Wen-tao, GUO Ming-zhang, LIU Hui-lin. Application of Synthetic Biology Based Whole-cell Biosensor Technology in the Rapid Detection of Food Safety [J]. Biotechnology Bulletin, 2023, 39(1): 137-149. |
| [14] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [15] | HU Hai-yang, YING Wan-qin, HE Jun, LV Zhi-xian, XIE Xiao-ping, DENG Zhong-liang. Establishment and Application of ERA Real-time Fluorescence Method for Rapid Detection of Mycoplasma pneumoniae [J]. Biotechnology Bulletin, 2022, 38(9): 264-270. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||