








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (11): 185-193.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0237
Previous Articles Next Articles
WANG Bo-ya( ), JIANG Yong, HUANG Yan, CAO Ying, HU Shang-lian(
), JIANG Yong, HUANG Yan, CAO Ying, HU Shang-lian( )
)
Received:2022-02-26
Online:2022-11-26
Published:2022-12-01
Contact:
HU Shang-lian
E-mail:287870877@qq.com;578404755@qq.com
WANG Bo-ya, JIANG Yong, HUANG Yan, CAO Ying, HU Shang-lian. Cloning and Functional Analysis of BeCesA4 in Bambusa emeiensis[J]. Biotechnology Bulletin, 2022, 38(11): 185-193.
| 引物名称Primer name | 序列Sequence(5'-3') | 用途Usage |
|---|---|---|
| BeCesA4F | TACTACTCGCGATACCCCATAG | 获得BeCesA4基因序列 Obtaining sequence of gene BeCesA4 |
| BeCesA4R | ATACTAACCTACGCCACCTCTC | |
| BeCesA4F(Kpn I) | GGGGTACCCCTACTACTCGCGATACCCCATAG | 构建植物表达载体 Construction of plant expression-vector |
| BeCesA4R(Xba I) | GCTCTAGAGCATACTAACCTACGCCACCTCTC | |
| BeCesA4-RTF | TCACCATCGGCAGCCACCT | 基因表达分析 Analysis of gene expression |
| BeCesA4-RTR | GCTTTTGCAGCAGCCTTTT | |
| Actin-RTF | AAACTGTAATGGTCCTCCCTCCG | 内参基因扩增 Amplification of internal reference |
| Actin-RTR | GCATCATCACAATCACTCTCCGA |
Table 1 Primer sequence
| 引物名称Primer name | 序列Sequence(5'-3') | 用途Usage |
|---|---|---|
| BeCesA4F | TACTACTCGCGATACCCCATAG | 获得BeCesA4基因序列 Obtaining sequence of gene BeCesA4 |
| BeCesA4R | ATACTAACCTACGCCACCTCTC | |
| BeCesA4F(Kpn I) | GGGGTACCCCTACTACTCGCGATACCCCATAG | 构建植物表达载体 Construction of plant expression-vector |
| BeCesA4R(Xba I) | GCTCTAGAGCATACTAACCTACGCCACCTCTC | |
| BeCesA4-RTF | TCACCATCGGCAGCCACCT | 基因表达分析 Analysis of gene expression |
| BeCesA4-RTR | GCTTTTGCAGCAGCCTTTT | |
| Actin-RTF | AAACTGTAATGGTCCTCCCTCCG | 内参基因扩增 Amplification of internal reference |
| Actin-RTR | GCATCATCACAATCACTCTCCGA |

Fig.2 BeCesA4 is homologous to AtCesA8,OsCesA4 and BdCesA8 in evolution A:Coding sequence of BeCesA4 and phylogenetic tree with CesA gene in species. The phylogenetic tree there are CesA family members from the species of Arabidopsis,Oryza sativa,Brachy podium distachyon,Populus tomentosa,Bambusa oldhamii and Phyllostachys heterocycla. B:Homologous sequences alignment of BeCesA4 with conserved domain in other species
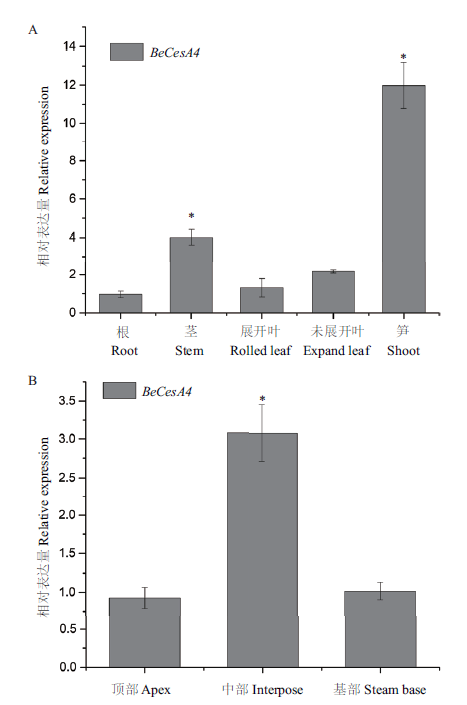
Fig.3 BeCesA4 notably expressed in fast-growing organs A:Expression of BeCesA4 in Bambusa emeiensis organs. B:BeCesA4 expressions differ in shoots at varied heights. Data represent means ± SD from 3 independent replicates. Asterisks indicate significant differences(*P<0.05)according to t test compared with other samples. The same below

Fig. 4 Biomass increment in BeCesA4 transgenic Populus tomentosa A:Expression analysis of BeCesA4 in transgenic populus. B:Height of transgenic populus and control. C:Growth of transgenic populus,bar length is 20 cm
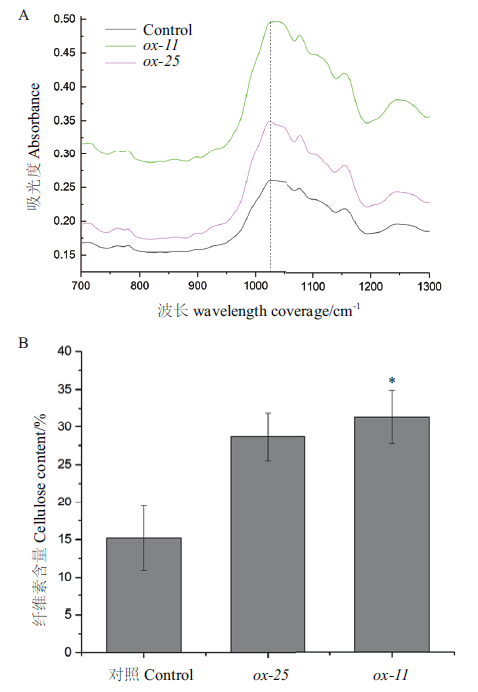
Fig.5 Increase of cellulose content in BeCesA4 transgenic Populus tomentosa A:Absorbance of BeCesA4 transgenic Populus plants and control. B:Cellulose content of BeCesA4 transgenic Populus plants and control
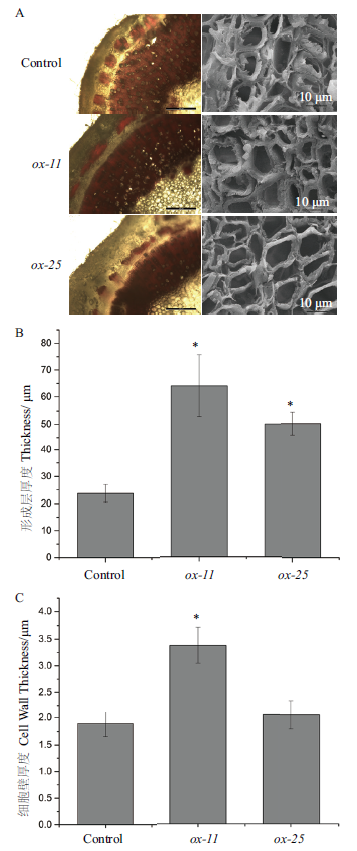
Fig.6 BeCesA4 participating in cell wall formation A-left:Phloroglucinol-hydrochloric staining of transgenic Populus tomentosa stem paraffin section. A-right:Scanning electron microscope image of transgenic Populus tomentosa stem paraffin sections. B:Cambium thickness of transgenic Populus tomentosa. C:Cell wall thickness of both transgenic Populus tomentosa and control
| [1] | China Paper Association. 中国造纸协会关于中国造纸工业2002年度报告[J]. 中华纸业, 2003, 24(5):6-9. |
| Association CP. A review on the development of China's paper industry in 2002[J]. China Pulp Pap Ind, 2003, 24(5):6-9. | |
| [2] |
Polko JK, Kieber JJ. The regulation of cellulose biosynthesis in plants[J]. Plant Cell, 2019, 31(2):282-296.
doi: 10.1105/tpc.18.00760 |
| [3] |
McFarlane HE, Döring A, Persson S. The cell biology of cellulose synthesis[J]. Annu Rev Plant Biol, 2014, 65:69-94.
doi: 10.1146/annurev-arplant-050213-040240 pmid: 24579997 |
| [4] | Pérez S, Samain D. Structure and engineering of celluloses[J]. Adv Carbohydr Chem Biochem, 2010, 64:25-116. |
| [5] | 高艳, 陈光辉, 陈秀娟, 等. 植物细胞壁纤维素生物合成的调控[J]. 生物技术通报, 2014(1):1-7. |
| Gao Y, Chen GH, Chen XJ, et al. Regulation of cellulose biosynthesis in plant cell wall[J]. Biotechnol Bull, 2014(1):1-7. | |
| [6] |
Wang LQ, Guo K, Li Y, et al. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice[J]. BMC Plant Biol, 2010, 10:282.
doi: 10.1186/1471-2229-10-282 URL |
| [7] |
Holland N, Holland D, Helentjaris T, et al. A comparative analysis of the plant cellulose synthase(CesA)gene family[J]. Plant Physiol, 2000, 123(4):1313-1324.
pmid: 10938350 |
| [8] |
Chen ZZ, Hong XH, Zhang HR, et al. Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis[J]. Plant J, 2005, 43(2):273-283.
doi: 10.1111/j.1365-313X.2005.02452.x URL |
| [9] |
Suzuki S, Li LG, Sun YH, et al. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa[J]. Plant Physiol, 2006, 142(3):1233-1245.
pmid: 16950861 |
| [10] |
Zhang SJ, Jiang ZX, Chen J, et al. The cellulose synthase(CesA)gene family in four Gossypium species:phylogenetics, sequence variation and gene expression in relation to fiber quality in Upland cotton[J]. Mol Genet Genomics, 2021, 296(2):355-368.
doi: 10.1007/s00438-020-01758-7 URL |
| [11] |
Xu WJ, Cheng H, Zhu SR, et al. Functional understanding of secondary cell wall cellulose synthases in Populus trichocarpa via the Cas9/gRNA-induced gene knockouts[J]. New Phytol, 2021, 231(4):1478-1495.
doi: 10.1111/nph.17338 URL |
| [12] |
Kumar M, Atanassov I, Turner S. Functional analysis of cellulose synthase(CESA)protein class specificity[J]. Plant Physiol, 2016, 173(2):970-983.
doi: 10.1104/pp.16.01642 URL |
| [13] |
Zhong RQ, Morrison WH 3rd, Freshour GD, et al. Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis[J]. Plant Physiol, 2003, 132(2):786-795.
pmid: 12805608 |
| [14] |
Park S, Ding SY. The N-terminal zinc finger of CELLULOSE SYNTHASE6 is critical in defining its functional properties by determining the level of homodimerization in Arabidopsis[J]. Plant J, 2020, 103(5):1826-1838.
doi: 10.1111/tpj.14870 URL |
| [15] |
Daras G, Templalexis D, Avgeri F, et al. Updating insights into the catalytic domain properties of plant Cellulose synthase(CesA)and Cellulose synthase-like(Csl)proteins[J]. Molecules, 2021, 26(14):4335.
doi: 10.3390/molecules26144335 URL |
| [16] |
Tanaka K, Murata K, Yamazaki M, et al. Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall[J]. Plant Physiol, 2003, 133(1):73-83.
pmid: 12970476 |
| [17] |
Li FC, Liu ST, Xu H, et al. A novel FC17/CESA4 mutation causes increased biomass saccharification and lodging resistance by remodeling cell wall in rice[J]. Biotechnol Biofuels, 2018, 11:298.
doi: 10.1186/s13068-018-1298-2 pmid: 30410573 |
| [18] |
Peng ZH, Lu Y, Li LB, et al. The draft genome of the fast-growing non-timber forest species moso bamboo(Phyllostachys heterocycla)[J]. Nat Genet, 2013, 45(4):456-461, 461e1-2.
doi: 10.1038/ng.2569 URL |
| [19] | Zhao HS, Gao ZM, Wang L, et al. Chromosome-level reference genome and alternative splicing atlas of moso bamboo(Phyllostachys edulis)[J]. GigaScience, 2018, 7(10):giy115. |
| [20] |
Guo ZH, Ma PF, Yang GQ, et al. Genome sequences provide insights into the reticulate origin and unique traits of woody bamboos[J]. Mol Plant, 2019, 12(10):1353-1365.
doi: 10.1016/j.molp.2019.05.009 URL |
| [21] |
Chen CY, Hsieh MH, Yang CC, et al. Analysis of the cellulose synthase genes associated with primary cell wall synthesis in Bambusa oldhamii[J]. Phytochemistry, 2010, 71(11/12):1270-1279.
doi: 10.1016/j.phytochem.2010.05.011 URL |
| [22] |
Huang HY, Cheng YS. Heterologous overexpression, purification and functional analysis of plant cellulose synthase from green bamboo[J]. Plant Methods, 2019, 15:80.
doi: 10.1186/s13007-019-0466-0 URL |
| [23] | 陈宇鹏, 曹颖, 胡尚连, 等. 基于高通量测序的慈竹笋转录组分析与基因功能注释[J]. 生物工程学报, 2016, 32(11):1610-1623. |
| Chen YP, Cao Y, Hu SL, et al. Transcriptome analysis and gene function annotation of Bambusa emeiensis shoots based on high-throughput sequencing technology[J]. Chin J Biotechnol, 2016, 32(11):1610-1623. | |
| [24] |
Mazarei M, Baxter HL, Li M, et al. Functional analysis of cellulose synthase CesA4 and CesA6 genes in switchgrass(Panicum virgatum)by overexpression and RNAi-mediated gene silencing[J]. Front Plant Sci, 2018, 9:1114.
doi: 10.3389/fpls.2018.01114 pmid: 30127793 |
| [25] |
Zhang BC, Deng LW, Qian Q, et al. A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice[J]. Plant Mol Biol, 2009, 71(4/5):509-524.
doi: 10.1007/s11103-009-9536-4 URL |
| [26] |
Fan CF, Feng SQ, Huang JF, et al. AtCesA8-driven OsSUS3 expression leads to largely enhanced biomass saccharification and lodging resistance by distinctively altering lignocellulose features in rice[J]. Biotechnol Biofuels, 2017, 10:221.
doi: 10.1186/s13068-017-0911-0 URL |
| [27] |
Li A, Xia T, Xu W, et al. An integrative analysis of four CESA isoforms specific for fiber cellulose production between Gossypium hirsutum and Gossypium barbadense[J]. Planta, 2013, 237(6):1585-1597.
doi: 10.1007/s00425-013-1868-2 URL |
| [28] |
Ye SW, Cai CY, Ren HB, et al. An efficient plant regeneration and transformation system of ma bamboo(Dendrocalamus latiflorus Munro)started from young shoot as explant[J]. Front Plant Sci, 2017, 8:1298.
doi: 10.3389/fpls.2017.01298 URL |
| [29] |
Ye SW, Chen G, Kohnen MV, et al. Robust CRISPR/Cas9 mediated genome editing and its application in manipulating plant height in the first generation of hexaploid Ma bamboo(Dendrocalamus latiflorus Munro)[J]. Plant Biotechnol J, 2020, 18(7):1501-1503.
doi: 10.1111/pbi.13320 URL |
| [30] | Huang BY, Zhuo RY, Fan HJ, et al. An efficient genetic transformation and CRISPR/Cas9-based genome editing system for moso bamboo(Phyllostachys edulis)[J]. Front Plant Sci, 2022, 13:822022. |
| [31] |
Hernández-Blanco C, Feng DX, Hu J, et al. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance[J]. Plant Cell, 2007, 19(3):890-903.
pmid: 17351116 |
| [32] |
Ramírez V, García-Andrade J, Vera P. Enhanced disease resistance to Botrytis cinerea in myb46 Arabidopsis plants is associated to an early down-regulation of CesA genes[J]. Plant Signal Behav, 2011, 6(6):911-913.
doi: 10.4161/psb.6.6.15354 URL |
| [33] |
Coolen S, van Pelt JA, van Wees SCM, et al. Mining the natural genetic variation in Arabidopsis thaliana for adaptation to sequential abiotic and biotic stresses[J]. Planta, 2019, 249(4):1087-1105.
doi: 10.1007/s00425-018-3065-9 URL |
| [34] | 徐梅卿, 戴玉成, 范少辉, 等. 中国竹类病害记述及其病原物分类地位(上)[J]. 林业科学研究, 2006, 19(6):692-699. |
| Xu MQ, Dai YC, Fan SH, et al. Records of bamboo diseases and the taxonomy of their pathogens in China(I)[J]. For Res, 2006, 19(6):692-699. |
| [1] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [2] | MA Yu-qian, SUN Dong-hui, YUE Hao-feng, XIN Jia-yu, LIU Ning, CAO Zhi-yan. Identification, Heterologous Expression and Functional Analysis of a GH61 Family Glycoside Hydrolase from Setosphaeria turcica with the Assisting Function in Degrading Cellulose [J]. Biotechnology Bulletin, 2023, 39(4): 124-135. |
| [3] | CHEN Nan-nan, WANG Chun-lai, JIANG Zhen-zhong, JIAO Peng, GUAN Shu-yan, MA Yi-yong. Genetic Transformation and Chilling Resistance Analysis of Maize ZmDHN15 Gene in Tobacco [J]. Biotechnology Bulletin, 2023, 39(4): 259-267. |
| [4] | NIU Xin, ZHANG Ying, WANG Mao-jun, LIU Wen-long, LU Fu-ping, LI Yu. Effects of Different Integration Sites on the Expression of Exogenous Alkaline Protease in Bacillus amyloliquefaciens [J]. Biotechnology Bulletin, 2022, 38(4): 253-260. |
| [5] | WANG Xiao-tao, ZOU Hang, WU Yi, XIANG Shen-wei, LV Hua, LIU Chao-lan, LIN Jia-fu, WANG Xin-rong, CHU Yi-wen, SONG Tao. Heterologous Expression and Enzymatic Properties Analysis of Novel β-agarase Aga2 from Paraglaciecola hydrolytica [J]. Biotechnology Bulletin, 2022, 38(11): 258-268. |
| [6] | ZHANG Tong-tong, ZHENG Deng-yu, WU Zhong-yi, ZHANG Zhong-bao, YU Rong. Functional Analysis of ZmNF-YB13 Responding to Drought and Salt Stress [J]. Biotechnology Bulletin, 2022, 38(10): 115-123. |
| [7] | LIAO Zhao-min, CAI Jun, LIN Jian-guo, DU Xin, WANG Chang-gao. Expression of Glucose Oxidase Gene from Aspergillus niger in Pichia pastoris and Optimization of Enzyme Production Conditions [J]. Biotechnology Bulletin, 2021, 37(6): 97-107. |
| [8] | LIU Shan, YE Wei, ZHU Mu-zi, LI Sai-ni, DENG Zhang-shuang, ZHANG Wei-min. Cloning,Expression and Characterization of a Novel Acyltransferase GPAT [J]. Biotechnology Bulletin, 2021, 37(11): 257-266. |
| [9] | ZHAO Hai-yan, SONG Chen-bin, LIU Zheng-ya, MA Xing-rong, SHANG Hui-hui, LI An-hua, GUAN Xian-jun, WANG Jian-she. Cloning,Recombinant Expression and Enzymatic Properties of α-Amylase Gene from Laceyella sp. [J]. Biotechnology Bulletin, 2020, 36(8): 23-33. |
| [10] | LI Wei-na, SHEN Dong-ling, ZHANG Yu-xing, LIU Xue-tong, IRBIS Chagan. Cloning and Enzymatic Identification of Thermo-tolerant and Endotype Alginate Lyase Gene from Mangrovibacterium sp. SH-52 [J]. Biotechnology Bulletin, 2020, 36(12): 82-90. |
| [11] | ZHAO Jiang-hua, FANG Huan, ZHANG Da-wei. Research Progress in Biosynthesis of Secondary Metabolites of Microorganisms [J]. Biotechnology Bulletin, 2020, 36(11): 141-147. |
| [12] | ZHANG Yu-wen, YUAN Hang, YU Jiang-yue, MA Xiao-xiao, SHI Chao-shuo, LI Yu. Screening of a Bacterial Strain Efficiently Degrading Feather Waste and Optimization of Its Expression Condition [J]. Biotechnology Bulletin, 2019, 35(9): 93-98. |
| [13] | GU Yuan, XUN Zi-qi, ZHENG Fei, TU Tao, YAO Bin, LUO Hui-ying. Identification and Functional Exploration of the Expansin Gene from Thermophilic Talaromyces leycettanus JCM12802 [J]. Biotechnology Bulletin, 2018, 34(10): 172-181. |
| [14] | CHEN Kun, YUAN Fei-yan, CHAI Hao-nan, LIU Huan, WANG Xing-ji, ZHANG Hui-tu, LU Fu-ping. Screening and Study of a New Promoter with Effective Expression of Alkaline Protease [J]. Biotechnology Bulletin, 2018, 34(1): 208-214. |
| [15] | ZHANG Duo-duo, ZHENG Fei, LUO Hui-ying, LI Zhong-yuan, LUO Xue-gang. Expression of α-Galactosidases from Thermoascus crustaceus JCM12803 in Pichia pastoris [J]. Biotechnology Bulletin, 2017, 33(6): 207-213. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||