








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (3): 276-284.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1138
Previous Articles Next Articles
LAN Xin-yue1,2( ), LIU Ning-ning2,3, ZHU Long-jiao2, CHEN Xu1,2, CHU Hua-shuo2, LI Xiang-yang4, DUAN Nuo5, XU Wen-tao1,2(
), LIU Ning-ning2,3, ZHU Long-jiao2, CHEN Xu1,2, CHU Hua-shuo2, LI Xiang-yang4, DUAN Nuo5, XU Wen-tao1,2( )
)
Received:2021-09-04
Online:2022-03-26
Published:2022-04-06
Contact:
XU Wen-tao
E-mail:lanxinyue1210@163.com;xuwentao@cau.edu.cn
LAN Xin-yue, LIU Ning-ning, ZHU Long-jiao, CHEN Xu, CHU Hua-shuo, LI Xiang-yang, DUAN Nuo, XU Wen-tao. Tetracycline Bivalent Aptamer Non-enzyme Label-free Sensor[J]. Biotechnology Bulletin, 2022, 38(3): 276-284.
| 名称Name | 序列Sequence(5'-3') | 碱基数Number of bases/nt |
|---|---|---|
| 四环素适配体 Tetracycline aptamer(TCA) | CGGTGGTG | 8 |
| 双价裁剪适配体 Bivalent shortened aptamer(TCSA) | CGGTGGTGCGGTGGTG | 16 |
| 间隔插入2T Intervening 2T(2T) | CGGTGGTGTTCGGTGGTG | 18 |
| 间隔插入2A Intervening 2A(2A) | CGGTGGTGAACGGTGGTG | 18 |
| 间隔插入2C Intervening 2C(2C) | 5CGGTGGTGCCCGGTGGTG | 18 |
| 间隔插入2G Intervening 2G(2G) | 5CGGTGGTGGGCGGTGGTG | 18 |
| 间隔插入1G Intervening 1G(1G) | CGGTGGTGGCGGTGGTG | 17 |
| 间隔插入3G Intervening 3G(3G) | CGGTGGTGGGGCGGTGGTG | 19 |
| 间隔插入4G Intervening 4G(4G) | CGGTGGTGGGGGCGGTGGTG | 20 |
| 间隔插入5G Intervening 5G(5G) | CGGTGGTGGGGGGCGGTGGTG | 21 |
Table 1 Sequences used in experiment
| 名称Name | 序列Sequence(5'-3') | 碱基数Number of bases/nt |
|---|---|---|
| 四环素适配体 Tetracycline aptamer(TCA) | CGGTGGTG | 8 |
| 双价裁剪适配体 Bivalent shortened aptamer(TCSA) | CGGTGGTGCGGTGGTG | 16 |
| 间隔插入2T Intervening 2T(2T) | CGGTGGTGTTCGGTGGTG | 18 |
| 间隔插入2A Intervening 2A(2A) | CGGTGGTGAACGGTGGTG | 18 |
| 间隔插入2C Intervening 2C(2C) | 5CGGTGGTGCCCGGTGGTG | 18 |
| 间隔插入2G Intervening 2G(2G) | 5CGGTGGTGGGCGGTGGTG | 18 |
| 间隔插入1G Intervening 1G(1G) | CGGTGGTGGCGGTGGTG | 17 |
| 间隔插入3G Intervening 3G(3G) | CGGTGGTGGGGCGGTGGTG | 19 |
| 间隔插入4G Intervening 4G(4G) | CGGTGGTGGGGGCGGTGGTG | 20 |
| 间隔插入5G Intervening 5G(5G) | CGGTGGTGGGGGGCGGTGGTG | 21 |
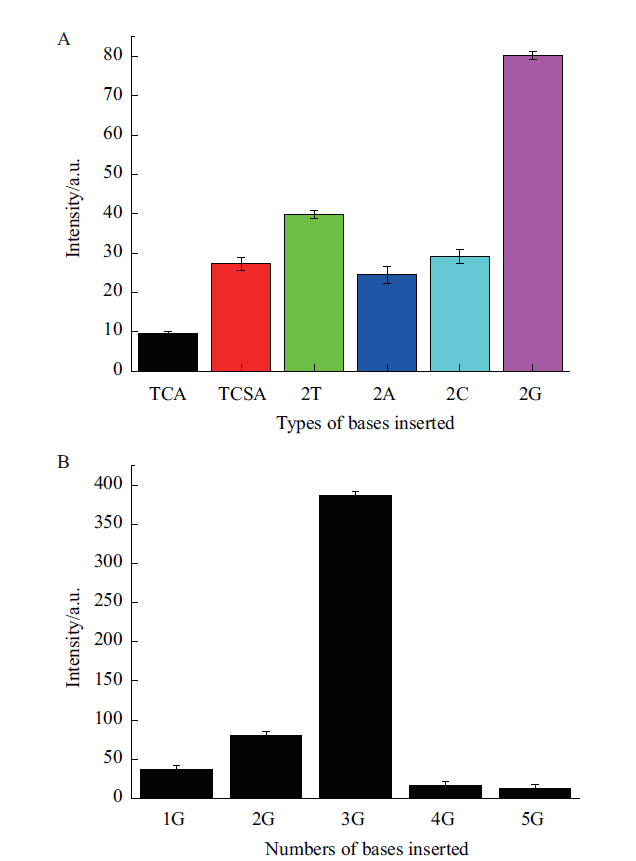
Fig. 2 Ability of bivalent aptamer spacer insert sequences to trigger ThT fluorescence A:Fluorescence intensity of different types of bases inserted at bivalent aptamer spacer. B:Fluorescence intensity of different numbers of G bases inserted at bivalent aptamer spacer
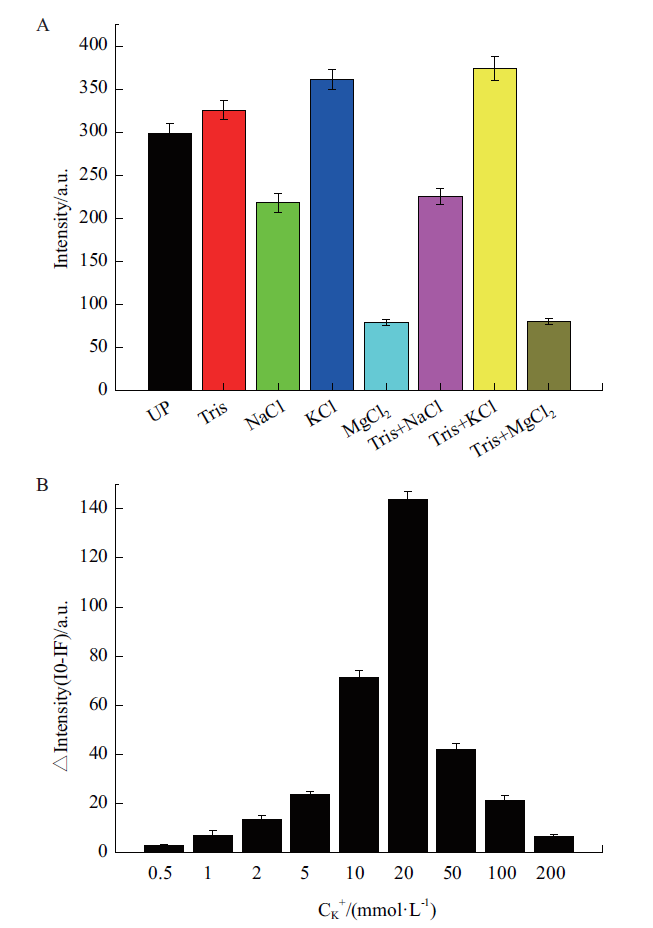
Fig. 7 Optimization of reaction system and ion concen-tration A:Fluorescence intensity of different buffer systems;Tris:20 mmol/L;NaCl:20 mmol/L;KCl:20 mmol/L;MgCl2:20 mmol/L. B:Changes of fluorescence intensity under different K+ concentration conditions before and after adding the target
| 添加浓度 Added concentration/(nmol·L-1) | △荧光强度 △ Fluorescence intensity/(I0-IF) | 回收浓度 Recycling concentration/(nmol·L-1) | 平均浓度 Average concentration/(nmol·L-1)+相对标准偏差 RSD | 回收率 Recovery rate/% | |||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | ||||
| 20 | 109.7 | 110.8 | 108.4 | 21.5 | 22.3 | 20.5 | 21.4±0.7 | 107.1666667 | |
| 100 | 155.9 | 156.0 | 155.2 | 103.9 | 104.1 | 101.4 | 103.1±1.2 | 103.1333334 | |
| 500 | 204.1 | 202.7 | 203.6 | 537.7 | 512.6 | 510.8 | 520.4±12.3 | 104.0733334 | |
Table 2 Standard addition and recovery of milk samples
| 添加浓度 Added concentration/(nmol·L-1) | △荧光强度 △ Fluorescence intensity/(I0-IF) | 回收浓度 Recycling concentration/(nmol·L-1) | 平均浓度 Average concentration/(nmol·L-1)+相对标准偏差 RSD | 回收率 Recovery rate/% | |||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | ||||
| 20 | 109.7 | 110.8 | 108.4 | 21.5 | 22.3 | 20.5 | 21.4±0.7 | 107.1666667 | |
| 100 | 155.9 | 156.0 | 155.2 | 103.9 | 104.1 | 101.4 | 103.1±1.2 | 103.1333334 | |
| 500 | 204.1 | 202.7 | 203.6 | 537.7 | 512.6 | 510.8 | 520.4±12.3 | 104.0733334 | |
| [1] |
Chatten LG, Krause SI. Colorimetric assay of tetracycline antibiotics[J]. J Pharm Sci, 1971, 60(1):107-110.
doi: 10.1002/jps.2600600121 URL |
| [2] |
Oka H, Ito Y, Matsumoto H. Chromatographic analysis of tetracycline antibiotics in foods[J]. J Chromatogr A, 2000, 882(1/2):109-133.
doi: 10.1016/S0021-9673(99)01316-3 URL |
| [3] |
Sun CY, Su RF, Bie JX, et al. Label-free fluorescent sensor based on aptamer and thiazole orange for the detection of tetracycline[J]. Dye Pigment, 2018, 149:867-875.
doi: 10.1016/j.dyepig.2017.11.031 URL |
| [4] | Ibarra IS, Rodriguez JA, Miranda JM, et al. Magnetic solid phase extraction based on phenyl silica adsorbent for the determination of tetracyclines in milk samples by capillary electrophoresis[J]. J Chromatogr A, 2011, 1218(16):2196-2202. |
| [5] | Zaitseva NV, Shur PZ, Atiskova NG, et al. Human health hazards associated with tetracycline drugs residues in food[J]. International Journal of Advanced Research, 2014, 2:488-495. |
| [6] | 于晓雯, 索全义. 畜禽粪便中四环素类抗生素的残留及危害[J]. 北方农业学报, 2018, 46(3):83-88. |
| Yu XW, Suo QY. Residues and hazards of tetracycline antibiotics in livestock and poultry manure[J]. J North Agric, 2018, 46(3):83-88. | |
| [7] |
Pastor-Navarro N, Morais S, Maquieira A, et al. Synjournal of haptens and development of a sensitive immunoassay for tetracycline residues. Application to honey samples[J]. Anal Chim Acta, 2007, 594(2):211-218.
pmid: 17586117 |
| [8] |
Samanidou VF, Nisyriou SA, Papadoyannis IN. Development and validation of an HPLC method for the determination of penicillin antibiotics residues in bovine muscle according to the European Union Decision 2002/657/EC[J]. J Sep Sci, 2007, 30(18):3193-3201.
pmid: 17960837 |
| [9] |
Kargin ID, Sokolova LS, Pirogov AV, et al. HPLC determination of tetracycline antibiotics in milk with post-column derivatization and fluorescence detection[J]. Inorg Mater, 2016, 52(14):1365-1369.
doi: 10.1134/S0020168516140065 URL |
| [10] |
Jiang T, Peng Z, Xie MP, et al. Rapid analysis of tetracycline in honey by microwave plasma torch mass spectrometry with ablation samples[J]. Anal Methods, 2020, 12(4):535-543.
doi: 10.1039/C9AY01887E URL |
| [11] |
Jing T, Gao XD, Wang P, et al. Determination of trace tetracycline antibiotics in foodstuffs by liquid chromatography-tandem mass spectrometry coupled with selective molecular-imprinted solid-phase extraction[J]. Anal Bioanal Chem, 2009, 393(8):2009-2018.
doi: 10.1007/s00216-009-2641-z pmid: 19214484 |
| [12] |
Zhang Y, Lu S, Liu W, et al. Preparation of anti-tetracycline antibodies and development of an indirect heterologous competitive enzyme-linked immunosorbent assay to detect residues of tetracycline in milk[J]. J Agric Food Chem, 2007, 55(2):211-218.
doi: 10.1021/jf062627s URL |
| [13] |
Tang Y, Huang X, Wang X, et al. G-quadruplex DNAzyme as peroxidase mimetic in a colorimetric biosensor for ultrasensitive and selective detection of trace tetracyclines in foods[J]. Food Chem, 2022, 366:130560.
doi: 10.1016/j.foodchem.2021.130560 URL |
| [14] |
Fu Q, Long C, Qin L, et al. Fluorescent and colorimetric dual-mode detection of tetracycline in wastewater based on heteroatoms-doped reduced state carbon dots[J]. Environ Pollut, 2021, 283:117109.
doi: 10.1016/j.envpol.2021.117109 URL |
| [15] |
Huy BT, Nghia NN, Lee YI. Highly sensitive colorimetric paper-based analytical device for the determination of tetracycline using green fluorescent carbon nitride nanoparticles[J]. Microchem J, 2020, 158:105151.
doi: 10.1016/j.microc.2020.105151 URL |
| [16] |
Shen L, Chen J, Li N, et al. Rapid colorimetric sensing of tetracycline antibiotics with in situ growth of gold nanoparticles[J]. Anal Chim Acta, 2014, 839:83-90.
doi: 10.1016/j.aca.2014.05.021 pmid: 25066722 |
| [17] |
Song JL, Huang MH, Lin XH, et al. Novel Fe-based metal-organic framework(MOF)modified carbon nanofiber as a highly selective and sensitive electrochemical sensor for tetracycline detection[J]. Chem Eng J, 2022, 427:130913.
doi: 10.1016/j.cej.2021.130913 URL |
| [18] |
Verma S, Ravichandiran V, Ranjan N. Beyond amyloid proteins:Thioflavin T in nucleic acid recognition[J]. Biochimie, 2021, 190:111-123.
doi: 10.1016/j.biochi.2021.06.003 URL |
| [19] |
Renaud de la Faverie A, Guédin A, Bedrat A, et al. Thioflavin T as a fluorescence light-up probe for G4 formation[J]. Nucleic Acids Res, 2014, 42(8):e65.
doi: 10.1093/nar/gku111 URL |
| [20] |
Amdursky N, Erez Y, Huppert D. Molecular rotors:what lies behind the high sensitivity of the thioflavin-T fluorescent marker[J]. Acc Chem Res, 2012, 45(9):1548-1557.
doi: 10.1021/ar300053p URL |
| [21] |
Mohanty J, Barooah N, Dhamodharan V, et al. Thioflavin T as an efficient inducer and selective fluorescent sensor for the human telomeric G-quadruplex DNA[J]. J Am Chem Soc, 2013, 135(1):367-376.
doi: 10.1021/ja309588h URL |
| [22] | Kim YS, Gu MB. Advances in aptamer screening and small molecule aptasensors[J]. Adv Biochem Eng Biotechnol, 2014, 140:29-67. |
| [23] |
Song KM, Lee S, Ban C. Aptamers and their biological applications[J]. Sensors:Basel, 2012, 12(1):612-631.
doi: 10.3390/s120100612 URL |
| [24] |
Wang T, Chen C, Larcher LM, et al. Three decades of nucleic acid aptamer technologies:Lessons learned, progress and opportunities on aptamer development[J]. Biotechnol Adv, 2019, 37(1):28-50.
doi: S0734-9750(18)30177-0 pmid: 30408510 |
| [25] | Kwon YS, Ahmad Raston NH, Gu MB. An ultra-sensitive colorimetric detection of tetracyclines using the shortest aptamer with highly enhanced affinity[J]. Chem Commun Camb Engl, 2014, 50(1):40-42. |
| [26] |
Ramezani M, Mohammad Danesh N, Lavaee P, et al. A novel colorimetric triple-helix molecular switch aptasensor for ultrasensitive detection of tetracycline[J]. Biosens Bioelectron, 2015, 70:181-187.
doi: 10.1016/j.bios.2015.03.040 pmid: 25814407 |
| [27] |
Zhang Z, Tao C, Yin J, et al. Enhancing the response rate of strand displacement-based electrochemical aptamer sensors using bivalent binding aptamer-cDNA probes[J]. Biosens Bioelectron, 2018, 103:39-44.
doi: 10.1016/j.bios.2017.12.027 URL |
| [28] | 沈永聪, 李守军, 杨林. 牛奶中抗生素残留检测技术进展[J]. 畜牧兽医科技信息, 2006(5):87-89. |
| Shen YC, Li SJ, Yang L. Progress in detection technology of antibiotic residues in milk[J]. Chin J Animal Husb Vet Med, 2006(5):87-89. | |
| [29] | 朱红. G-quadruplex结构和稳定性的分子模拟研究[D]. 合肥:中国科学技术大学, 2015. |
| Zhu H. Structural Properties and Stability of G-quadruplex Studied by Molecular Simulations[D]. Hefei:University of Science and Technoloay of China, 2015. | |
| [30] |
Choudhury SD, Mohanty J, Pal H, et al. Cooperative metal ion binding to a cucurbit[7]uril-thioflavin T complex:demonstration of a stimulus-responsive fluorescent supramolecular capsule[J]. J Am Chem Soc, 2010, 132(4):1395-1401.
doi: 10.1021/ja908795y URL |
| [31] |
Stsiapura VI, Maskevich AA, Kuzmitsky VA, et al. Thioflavin T as a molecular rotor:fluorescent properties of thioflavin T in solvents with different viscosity[J]. J Phys Chem B, 2008, 112(49):15893-15902.
doi: 10.1021/jp805822c URL |
| [32] |
Xue WF, Hellewell AL, Gosal WS, et al. Fibril fragmentation enhances amyloid cytotoxicity[J]. J Biol Chem, 2009, 284(49):34272-34282.
doi: 10.1074/jbc.M109.049809 URL |
| [33] |
Biancardi A, Biver T, Burgalassi A, et al. Mechanistic aspects of thioflavin-T self-aggregation and DNA binding:evidence for dimer attack on DNA grooves[J]. Phys Chem Chem Phys, 2014, 16(37):20061-20072.
doi: 10.1039/c4cp02838d pmid: 25130260 |
| [1] | LI Hui-jie, DONG Lian-hua, CHEN Gui-fang, LIU Si-yuan, YANG Jia-yi, YANG Jing-ya. Establishment of Droplet Digital PCR Assay for Quantitative Detection of Pseudomonas cocovenenans in Foods [J]. Biotechnology Bulletin, 2023, 39(1): 127-136. |
| [2] | LIU Ning-ning, WANG Xin-xin, LAN Xin-yue, CHU Hua-shuo, CHEN Xu, CHANG Shi-min, LI Teng-fei, XU Wen-tao. G-Triplex Visualization Nucleic Acid Sensor for the Detection of Tetracycline [J]. Biotechnology Bulletin, 2022, 38(10): 106-114. |
| [3] | FENG Min, LI Shu-ting, ZHANG Yang-zi, SU Yuan, ZHU Long-jiao, CAO Ji-juan, LIU Hai-yan, XU Wen-tao. Development of a Innovative Fluorescent Quantitative PCR Method for Salmonella Based on Fluorescent Self-quenching Primers [J]. Biotechnology Bulletin, 2021, 37(11): 285-292. |
| [4] | WU Xue-ling, ZHOU Xiang-yu, WU Xiao-yan, LUO Kui, GU Yi-chao, ZHOU Han, LIAO Wan-qing, ZENG Wei-min. Construction of Tetracycline-degrading Bacterial Co-culture System and Community Analysis of Wastewater Remediation [J]. Biotechnology Bulletin, 2020, 36(10): 116-126. |
| [5] | XIAO Bing, LUO Yun-bo, HUANG Kun-lun, ZHANG Yuan, XU Wen-tao. Research Progress in the Quantitative and Unitive Detecting Technologies Based on Functional Nucleic Acid and Labeled Fluorescence [J]. Biotechnology Bulletin, 2019, 35(7): 213-221. |
| [6] | XIAO Bing, LIU Bang, LUO Yun-bo, HUANG Kun-lun, ZHANG Yuan, LI Xia-ying, ZHANG Xiu-jie, XU Wen-tao, ZHOU Xiang. Research Progress in Quantitative and Unitive Detecting Technologies of Functional Nucleic Acid and Label-Free Fluorescence [J]. Biotechnology Bulletin, 2019, 35(3): 194-202. |
| [7] | ZHANG Yong-li, YUAN Wei, YANG Qing-xiang. Effects of Nano-TiO2 and Tetracycline Exposure on Drug-resistant Escherichia coli [J]. Biotechnology Bulletin, 2019, 35(11): 124-131. |
| [8] | GAO Zhi-qiang, WANG Lin, PU Jing, YIN Yi, ZHANG Wei, ZHAO Xiang-peng, YAO Zhen-yu. Duplex Real-time PCR Methods for Quantitative Detection of Bovine Derived Materials in Animal Products [J]. Biotechnology Bulletin, 2018, 34(9): 190-194. |
| [9] | WU Xue-ling, WU Xiao-yan, LI Jiao-kun, SHEN Li, YU Run-lan, ZENG Wei-min. Isolation and Degradation Characteristics of a Efficient Tetracycline-degrading Strain [J]. Biotechnology Bulletin, 2018, 34(5): 172-178. |
| [10] | YU Xiao-jun, CAO Shao-yu, DONG Yu-mei, BI Bao-liang, ZHANG Ying-hua, XU Jun-qiang. Activity Detection of Chloroplast Promoter Based on Tetracycline Regulatory System in Escherichia coli [J]. Biotechnology Bulletin, 2017, 33(2): 149-154. |
| [11] | Zhang Xinyang, Cai Tingjing, Xu Xuping. Isolation and Identification of A Tetracycline-degrading Bacterium and Optimizing Condition for Tetracycline Degradation [J]. Biotechnology Bulletin, 2015, 31(1): 173-180. |
| [12] | Bai Helu, Liu Rui, Zhu Naishuo . Cell Model Construction of Regulated Hepatitis B Virus X Gene Expression [J]. Biotechnology Bulletin, 2013, 0(2): 140-146. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||