








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (4): 163-173.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1272
Previous Articles Next Articles
MA Xin-xin( ), XU Yang, ZHAO Huan-huan, HUO Zhao-yan, WANG Shu-bin, ZHONG Feng-lin(
), XU Yang, ZHAO Huan-huan, HUO Zhao-yan, WANG Shu-bin, ZHONG Feng-lin( )
)
Received:2021-10-09
Online:2022-04-26
Published:2022-05-06
Contact:
ZHONG Feng-lin
E-mail:1126691073@qq.com;zhong591@fafu.edu.cn
MA Xin-xin, XU Yang, ZHAO Huan-huan, HUO Zhao-yan, WANG Shu-bin, ZHONG Feng-lin. Identification of Tomato 4CL Gene Family and Expression Analysis Under Nitrogen Treatment[J]. Biotechnology Bulletin, 2022, 38(4): 163-173.
| 类型 Type | 基因ID Gene ID | 基因名称Gene name | 正向引物Forward(5'-3') | 反向引物Reverse(5'-3') |
|---|---|---|---|---|
| RT-qPCR | Solyc03g097030 | Sl4CL01 | TTTGAGGAGAGGACATGTATTATCG | GACGATGCTAGAATGGTAAA |
| Solyc03g117870 | Sl4CL02 | ATGTTGACACACAAAGGCT | ATTGCTGCTCCGACTCTC | |
| Solyc06g068650 | Sl4CL03 | TGTCTTCCTTGGTGCGTC | GCATAATCCTTCACCTTGTTC | |
| Solyc08g076300 | Sl4CL04 | TAAGTCAGCAAGATACAGCG | GAGAGAAAGCCCATACACAT | |
| Solyc11g069050 | Sl4CL05 | TCAACCTATGAGTATCTTTGCTGC | AAAACCGTTGTGCCGTGC | |
| Solyc12g042460 | Sl4CL06 | AGTAAGGATAGTCATTTCTGGG | GCTAAGCACATTGTTATAGGTG | |
| Actin | GTCCTCTTCCAGCCATCCAT | ACCACTGAGCACAATGTTACCG | ||
| RT-PCR | Solyc06g068650 | Sl4CL03 | ATGACAAATGCAACGATGGAG | ATTTGGA TATCCAGCAG CT |
| pCAMBIA1302 | GTGGATTGATGTGATATCTCC | CTGACAGAAAATTTGTGCCC |
Table 1 Primers for PCR
| 类型 Type | 基因ID Gene ID | 基因名称Gene name | 正向引物Forward(5'-3') | 反向引物Reverse(5'-3') |
|---|---|---|---|---|
| RT-qPCR | Solyc03g097030 | Sl4CL01 | TTTGAGGAGAGGACATGTATTATCG | GACGATGCTAGAATGGTAAA |
| Solyc03g117870 | Sl4CL02 | ATGTTGACACACAAAGGCT | ATTGCTGCTCCGACTCTC | |
| Solyc06g068650 | Sl4CL03 | TGTCTTCCTTGGTGCGTC | GCATAATCCTTCACCTTGTTC | |
| Solyc08g076300 | Sl4CL04 | TAAGTCAGCAAGATACAGCG | GAGAGAAAGCCCATACACAT | |
| Solyc11g069050 | Sl4CL05 | TCAACCTATGAGTATCTTTGCTGC | AAAACCGTTGTGCCGTGC | |
| Solyc12g042460 | Sl4CL06 | AGTAAGGATAGTCATTTCTGGG | GCTAAGCACATTGTTATAGGTG | |
| Actin | GTCCTCTTCCAGCCATCCAT | ACCACTGAGCACAATGTTACCG | ||
| RT-PCR | Solyc06g068650 | Sl4CL03 | ATGACAAATGCAACGATGGAG | ATTTGGA TATCCAGCAG CT |
| pCAMBIA1302 | GTGGATTGATGTGATATCTCC | CTGACAGAAAATTTGTGCCC |
| 基因名称Gene name | 氨基酸个数 Number of amino acids | 分子量 Molecular weight/kD | 等电点 Isoelectric point/pI | 不稳定指数Instability index | 总平均亲水系数Grand average of hydropathicity | 原子总数Total number of atoms | 亚细胞定位Subcellular localization |
|---|---|---|---|---|---|---|---|
| Sl4CL01 | 570 | 62080.54 | 5.34 | 34.92 | 0.106 | 8820 | Peroxisome Chloroplast |
| Sl4CL02 | 699 | 77723.64 | 5.64 | 37.7 | -0.126 | 10926 | Peroxisome |
| Sl4CL03 | 560 | 61700.2 | 5.49 | 33.69 | -0.028 | 8735 | Peroxisome |
| Sl4CL04 | 957 | 102255.02 | 5.45 | 31.47 | 0.476 | 14550 | Peroxisome |
| Sl4CL05 | 538 | 58930.46 | 8.49 | 30.69 | 0.07 | 8369 | Peroxisome |
| Sl4CL06 | 548 | 60394.02 | 5.9 | 32.36 | 0.045 | 8590 | Peroxisome |
Table 2 Physico-chemical properties of 4CL family proteins in tomato(Solanum lycopersicum)
| 基因名称Gene name | 氨基酸个数 Number of amino acids | 分子量 Molecular weight/kD | 等电点 Isoelectric point/pI | 不稳定指数Instability index | 总平均亲水系数Grand average of hydropathicity | 原子总数Total number of atoms | 亚细胞定位Subcellular localization |
|---|---|---|---|---|---|---|---|
| Sl4CL01 | 570 | 62080.54 | 5.34 | 34.92 | 0.106 | 8820 | Peroxisome Chloroplast |
| Sl4CL02 | 699 | 77723.64 | 5.64 | 37.7 | -0.126 | 10926 | Peroxisome |
| Sl4CL03 | 560 | 61700.2 | 5.49 | 33.69 | -0.028 | 8735 | Peroxisome |
| Sl4CL04 | 957 | 102255.02 | 5.45 | 31.47 | 0.476 | 14550 | Peroxisome |
| Sl4CL05 | 538 | 58930.46 | 8.49 | 30.69 | 0.07 | 8369 | Peroxisome |
| Sl4CL06 | 548 | 60394.02 | 5.9 | 32.36 | 0.045 | 8590 | Peroxisome |
| 基因名称 Gene name | α-螺旋α-helix/% | 延伸链 Extended strand/% | β-转角 β-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|
| Sl4CL01 | 32.28 | 18.6 | 6.84 | 42.28 |
| Sl4CL02 | 33.19 | 18.6 | 9.01 | 39.2 |
| Sl4CL03 | 30.89 | 20.71 | 7.5 | 40.89 |
| Sl4CL04 | 41.69 | 15.57 | 7.94 | 34.8 |
| Sl4CL05 | 30.67 | 20.07 | 8.36 | 40.89 |
| Sl4CL06 | 31.93 | 19.53 | 8.58 | 39.96 |
Table 3 Secondary structure of tomato 4CL gene family
| 基因名称 Gene name | α-螺旋α-helix/% | 延伸链 Extended strand/% | β-转角 β-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|
| Sl4CL01 | 32.28 | 18.6 | 6.84 | 42.28 |
| Sl4CL02 | 33.19 | 18.6 | 9.01 | 39.2 |
| Sl4CL03 | 30.89 | 20.71 | 7.5 | 40.89 |
| Sl4CL04 | 41.69 | 15.57 | 7.94 | 34.8 |
| Sl4CL05 | 30.67 | 20.07 | 8.36 | 40.89 |
| Sl4CL06 | 31.93 | 19.53 | 8.58 | 39.96 |
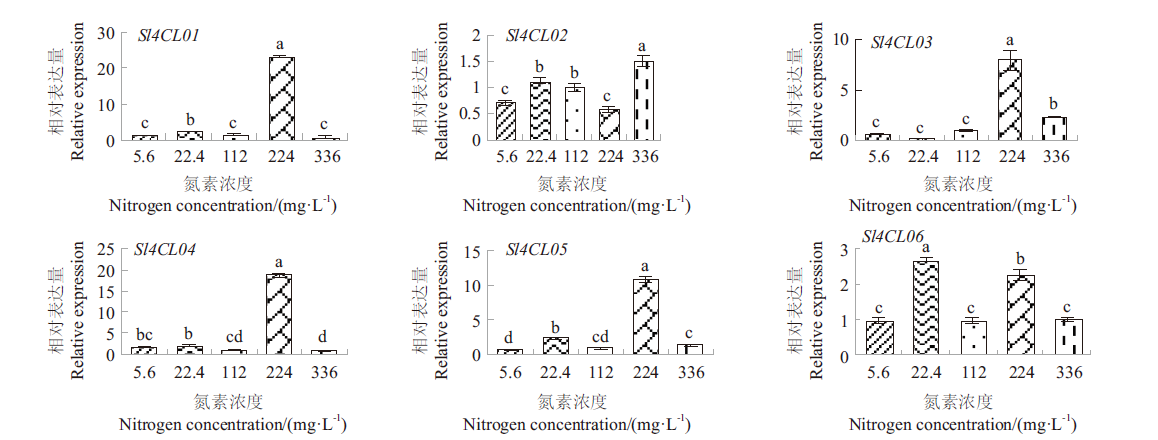
Fig.6 Expression patterns of tomato 4CL genes under different concentrations of nitrogen Histograms with different letters indicate significant difference at the 0.05 level
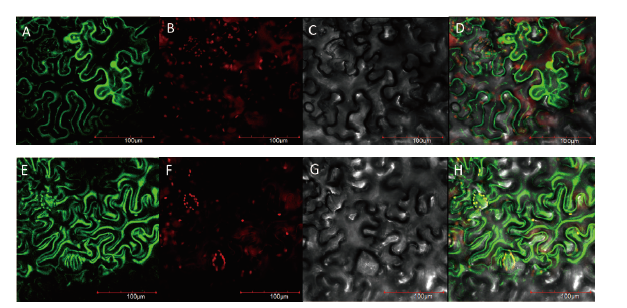
Fig.8 Subcellular localization of Sl4CL03 in Nicotiana tabacum leaf cells A-D:pCAMBIA1302 empty vector fluorescence,chloroplast fluorescence,bright,and merged.E-H:Sl4CL03 protein fluorescence,chloroplast fluorescence,bright,and merged
| [1] | 杜红豆. 氮素对飞机草(Eupatorium odoratum)4CL 酶活性和基因表达影响的研究[D]. 哈尔滨:哈尔滨师范大学, 2012. |
| Du HD. Influence of nitrogen on activity of 4-coumarate:coenzyme a ligase(4CL)and its gene expression in Eupatorium odoratum[D]. Harbin:Harbin Normal University, 2012. | |
| [2] | 李晓瑞. 干旱胁迫下小麦苯丙烷类代谢及MYB转录因子TaMpc1-D4响应机制研究[D]. 杨凌:西北农林科技大学, 2020. |
| Li XR. The mechanisms of the phenylpropanoid pathway and MYB transcription factor TaMpc1-D4 in responses to drought stress in wheat[D]. Yangling:Northwest A&F University, 2020. | |
| [3] | 常胜合, 孙威, 许桂莺, 等. 利用4-香豆酸:CoA连接酶基因Mu4CL15提高香蕉植株的倒伏抗性[J]. 分子植物育种, 2017, 15(12):4905-4911. |
| Chang SH, Sun W, Xu GY, et al. Enhancing lodging resistance of banana plant by transforming 4-coumarate:CoA ligase gene Mu4CL15[J]. Mol Plant Breed, 2017, 15(12):4905-4911. | |
| [4] |
Sun SC, Xiong XP, Zhang XL, et al. Correction to:Characterization of the Gh4CL gene family reveals a role of Gh4CL7 in drought tolerance[J]. BMC Plant Biol, 2021, 21(1):65.
doi: 10.1186/s12870-021-02834-9 URL |
| [5] | Zhang MY, Wang DJ, Gao XX, et al. Exogenous caffeic acid and epicatechin enhance resistance against Botrytis cinerea through activation of the phenylpropanoid pathway in apples[J]. Sci Hortic, 2020, 268:109348. |
| [6] | Zhang CH, Ma T, Luo WC, et al. Identification of 4CL genes in desert poplars and their changes in expression in response to salt stress[J]. Genes:Basel, 2015, 6(3):901-917. |
| [7] |
Wei HY, Rao GD, Wang YK, et al. Cloning and analysis of a new 4CL-like gene in Populus tomentosa[J]. For Sci Pract, 2013, 15(2):98-104.
doi: 10.1007/s11632-013-0204-z URL |
| [8] | Liu XY, Cui XM, Ji DC, et al. Luteolin-induced activation of the phenylpropanoid metabolic pathway contributes to quality maintenance and disease resistance of sweet cherry[J]. Food Chem, 2021, 342:128309. |
| [9] |
Dong NQ, Lin HX. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions[J]. J Integr Plant Biol, 2021, 63(1):180-209.
doi: 10.1111/jipb.13054 URL |
| [10] |
Lavhale SG, Kalunke RM, Giri AP. Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants[J]. Planta, 2018, 248(5):1063-1078.
doi: 10.1007/s00425-018-2965-z URL |
| [11] | Li SS, Li Y, Sun LL, et al. Identification and expression analysis of 4-Coumarate:Coenzyme A ligase gene family in Dryopteris Fragrans[J]. Cell Mol Biol:Noisy-Le-Grand, 2015, 61(4):25-33. |
| [12] |
Badhan S, Ball AS, Mantri N. First report of CRISPR/Cas9 mediated DNA-free editing of 4CL and RVE7 genes in chickpea protoplasts[J]. Int J Mol Sci, 2021, 22(1):396.
doi: 10.3390/ijms22010396 URL |
| [13] |
王惠, 张顺斌, 金贺, 等. 4-香豆酸辅酶A连接酶响应大豆孢囊线虫胁迫的潜在功能[J]. 生物技术通报, 2021, 37(7):71-80.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-0378 |
| Wang H, Zhang SB, Jin H, et al. Potential function of 4-coumaric acid-CoA ligase in response to soybean cyst nematode stress[J]. Biotechnol Bull, 2021, 37(7):71-80. | |
| [14] | 杨克彬, 单雪萌, 史晶晶, 等. 毛竹4-香豆酸辅酶A连接酶基因家族鉴定及表达分析[J]. 核农学报, 2021, 35(1):72-82. |
| Yang KB, Shan XM, Shi JJ, et al. Identification and expression analysis of 4CL gene family in Phyllostachys edulis[J]. J Nucl Agric Sci, 2021, 35(1):72-82. | |
| [15] | 洪平静, 徐小萍, 王静宇, 等. 龙眼体胚发生早期4CL基因家族鉴定与功能分析[J]. 热带作物学报, 2021, 42(4):909-919. |
| Hong PJ, Xu XP, Wang JY, et al. Identification and functional analysis of 4CL gene family during early somatic embryogenesis in Dimocarpus longan lour[J]. Chin J Trop Crop, 2021, 42(4):909-919. | |
| [16] | 唐映红. 苎麻4CL和CCR基因家族成员的克隆、表达及功能研究[D]. 长沙:湖南农业大学, 2018. |
| Tang YH. Study on the cloning, expression and functional of Boehmeria nivea 4CL and CCR gene family members[D]. Changsha:Hunan Agricultural University, 2018. | |
| [17] | 申晚霞, 王志彬, 薛杨, 等. 柑橘4CL基因家族的结构及其功能分析[J]. 园艺学报, 2019, 46(6):1068-1078. |
| Shen WX, Wang ZB, Xue Y, et al. Characterization of 4-coumarate:CoA ligase(4CL)gene family in Citrus[J]. Acta Hortic Sin, 2019, 46(6):1068-1078. | |
| [18] | 陈夏晔, 彭宣翔. 普通烟草Nt4CL基因家族的生物信息学分析[J]. 贵州农业科学, 2018, 46(8):11-15. |
| Chen XY, Peng XX. Bioinformatics analysis on tobacco Nt4CL gene family[J]. Guizhou Agric Sci, 2018, 46(8):11-15. | |
| [19] | 曹运鹏, 方志, 李姝妹, 等. 砀山酥梨4CL基因家族的全基因组鉴定与分析[J]. 遗传, 2015, 37(7):711-719. |
| Cao YP, Fang Z, Li SM, et al. Genome-wide identification and analyses of 4CL gene families in Pyrus bretschneideri Rehd[J]. Hereditas, 2015, 37(7):711-719. | |
| [20] |
Meng J, Li CN, Zhao ML, et al. Lignin biosynjournal regulated by the antisense 4CL gene in alfalfa[J]. Czech J Genet Plant Breed, 2018, 54(No. 1):26-29.
doi: 10.17221/23/2017-CJGPB URL |
| [21] | 孙士超. 绿色棉纤维转录组分析及4CL基因家族的鉴定[D]. 石河子:石河子大学, 2020. |
| Sun SC. Transcriptome analysis of green cotton fiber and identification of 4CL gene family[D]. Shihezi:Shihezi University, 2020. | |
| [22] | 杨婷, 潘翔, 饶国栋, 等. 植物4CL基因家族结构功能与表达特性研究进展[J]. 成都大学学报:自然科学版, 2011, 30(1):4-7. |
| Yang T, Pan X, Rao GD, et al. Research progress in structural function and expression characteristic of 4CL’s gene family in plants[J]. J Chengdu Univ:Nat Sci Ed, 2011, 30(1):4-7. | |
| [23] |
田晓明, 颜立红, 向光锋, 等. 植物4香豆酸:辅酶A连接酶研究进展[J]. 生物技术通报, 2017, 33(4):19-26.
doi: 10.13560/j.cnki.biotech.bull.1985.2017.04.003 |
| Tian XM, Yan LH, Xiang GF, et al. Research progress on 4-coumarate:coenzyme A ligase(4CL)in plants[J]. Biotechnol Bull, 2017, 33(4):19-26. | |
| [24] | 乔中全, 王晓明, 曾慧杰, 等. 灰毡毛忍冬Lm4CL基因克隆及表达分析[J]. 中南林业科技大学学报, 2021, 41(5):122-132. |
| Qiao ZQ, Wang XM, Zeng HJ, et al. Clone and expression analysis of Lm4CL in Lonicera macranthoides Hand-Mazz[J]. J Central South Univ For Technol, 2021, 41(5):122-132. | |
| [25] |
Sun H, Guo K, Feng S, et al. Positive selection drives adaptive diversification of the 4-coumarate:CoA ligase(4CL)gene in angiosperms[J]. Ecol Evol, 2015, 5(16):3413-3420.
doi: 10.1002/ece3.1613 URL |
| [26] | 汤崴, 白云赫, 胡雨晴, 等. 葡萄木质素相关基因PAL、4CL和CAD特征鉴定及其应答赤霉素在果实发育过程中的作用[J]. 南京农业大学学报, 2019, 42(3):430-439. |
| Tang W, Bai YH, Hu YQ, et al. Characterization of grape lignin-related genes PAL, 4CL, CAD and the role of GA in the berry development[J]. J Nanjing Agric Univ, 2019, 42(3):430-439. | |
| [27] | 王星淇, 孙雪丽, 车婧如, 等. 两个非洲菊4CL基因的克隆及表达[J]. 应用与环境生物学报, 2020, 26(2):272-279. |
| Wang XQ, Sun XL, Che JR, et al. Cloning and expression analysis of two 4CL genes in Gerbera[J]. Chin J Appl Environ Biol, 2020, 26(2):272-279. | |
| [28] |
Park JJ, Yoo CG, Flanagan A, et al. Defined Tetra-allelic gene disruption of the 4-coumarate:coenzyme A ligase 1(Pv4CL1)gene by CRISPR/Cas9 in switchgrass results in lignin reduction and improved sugar release[J]. Biotechnol Biofuels, 2017, 10:284.
doi: 10.1186/s13068-017-0972-0 URL |
| [29] | Muna Alariqi. 陆地棉基因GhLDOX和Gh4CL3的功能解析[D]. 武汉:华中农业大学, 2020. |
| Muna Alariqi. Functional studies of GhLDOX & Gh4CL3 genes in upland cotton Gossypium hirsutum[D]. Wuhan:Huazhong Agricultural University, 2020. | |
| [30] | 熊显鹏. Gh4CL30和GhWRKY70D13在棉花抗黄萎病中的功能研究[D]. 石河子:石河子大学, 2020. |
| Xiong XP. Functional analysis of Gh4CL30 and GhWRKY70D13 in Cotton Resistance to Verticillium Wilt[D]. Shihezi:Shihezi University, 2020. | |
| [31] |
Kienow L, Schneider K, Bartsch M, et al. Jasmonates meet fatty acids:functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana[J]. J Exp Bot, 2008, 59(2):403-419.
doi: 10.1093/jxb/erm325 pmid: 18267944 |
| [1] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [2] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [3] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [4] | SHI Jian-lei, ZAI Wen-shan, SU Shi-wen, FU Cun-nian, XIONG Zi-li. Identification and Expression Analysis of miRNA Related to Bacterial Wilt Resistance in Tomato [J]. Biotechnology Bulletin, 2023, 39(5): 233-242. |
| [5] | CHE Yong-mei, GUO Yan-ping, LIU Guang-chao, YE Qing, LI Ya-hua, ZHAO Fang-gui, LIU Xin. Isolation and Identification of Bacterial Strain C8 and B4 and Their Halotolerant Growth-promoting Effects and Mechanisms [J]. Biotechnology Bulletin, 2023, 39(5): 276-285. |
| [6] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [7] | HU Ming-yue, YANG Yu, GUO Yang-dong, ZHANG Xi-chun. Functional Analysis of SlMYB96 Gene in Tomato Under Cold Stress [J]. Biotechnology Bulletin, 2023, 39(4): 236-245. |
| [8] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [9] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [10] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [11] | SHEN Yun-xin, SHI Zhu-feng, ZHOU Xu-dong, LI Ming-gang, ZHANG Qing, FENG Lu-yao, CHEN Qi-bin, YANG Pei-wen. Isolation, Identification and Bio-activity of Three Bacillus Strains with Biocontrol Function [J]. Biotechnology Bulletin, 2023, 39(3): 267-277. |
| [12] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| [13] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [14] | GE Wen-dong, WANG Teng-hui, MA Tian-yi, FAN Zhen-yu, WANG Yu-shu. Genome-wide Identification of the PRX Gene Family in Cabbage(Brassica oleracea L. var. capitata)and Expression Analysis Under Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 252-260. |
| [15] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||