








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (6): 147-156.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1174
Previous Articles Next Articles
YANG Jia-bao1( ), ZHOU Zhi-ming1, ZHANG Zhan2, FENG Li1, SUN Li1(
), ZHOU Zhi-ming1, ZHANG Zhan2, FENG Li1, SUN Li1( )
)
Received:2021-09-13
Online:2022-06-26
Published:2022-07-11
Contact:
SUN Li
E-mail:2516040371@qq.com;lis@shzu.edu.cn
YANG Jia-bao, ZHOU Zhi-ming, ZHANG Zhan, FENG Li, SUN Li. Cloning,Expression of Helianthus annuus HaLACS1 Gene and Identification of Its Functional Complementation in Saccharomyces cerevisiae[J]. Biotechnology Bulletin, 2022, 38(6): 147-156.
| 引物名称 Primer | 引物序列 Primer sequence(5'-3') | 功能 Function |
|---|---|---|
| L1F | CCCAAGCTTATGGATAAGCTG- CAGTTTGC | HaLACS1克隆及酵母表达载体构建 |
| L1R | CCGCTCGAGTTAAGACTTGTT- CCCGCCTA | |
| qF | GAGCTCGAAAACGGAACGTG | qRT-PCR |
| qR | ATTGCAAGCCTCCATCGCTA | |
| PF | CGGACTAGTATGGATAAGCT- GCAGTTT | pAN580-HaLACS1-eGFP载体构建 |
| PR | CATGCCATGGAGACTTGTTC- CCGCCTA | |
| 18S-F | CTACCACATCCAAGGAAGG- CAG | qRT-PCR |
| 18S-R | CGACAGAAGGGACGAGTA- AACC |
Table 1 Information of primers used in this study
| 引物名称 Primer | 引物序列 Primer sequence(5'-3') | 功能 Function |
|---|---|---|
| L1F | CCCAAGCTTATGGATAAGCTG- CAGTTTGC | HaLACS1克隆及酵母表达载体构建 |
| L1R | CCGCTCGAGTTAAGACTTGTT- CCCGCCTA | |
| qF | GAGCTCGAAAACGGAACGTG | qRT-PCR |
| qR | ATTGCAAGCCTCCATCGCTA | |
| PF | CGGACTAGTATGGATAAGCT- GCAGTTT | pAN580-HaLACS1-eGFP载体构建 |
| PR | CATGCCATGGAGACTTGTTC- CCGCCTA | |
| 18S-F | CTACCACATCCAAGGAAGG- CAG | qRT-PCR |
| 18S-R | CGACAGAAGGGACGAGTA- AACC |

Fig. 2 Multiple alignment results of LACS1 proteins from different plants At:Arabidopsis thaliana. Ha:H. annuus L. Ls:Lactuca sativa. Bn:Brassica napus. Zm:Zea mays. Os:Oryza sativa
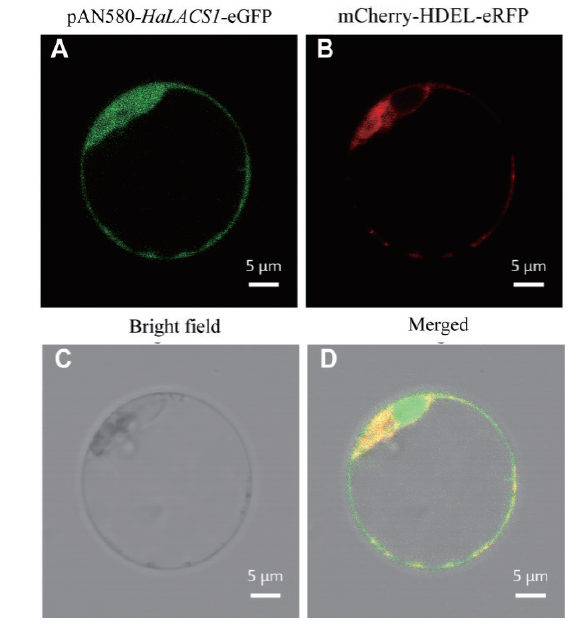
Fig. 5 Subcellular localization of the HaLACS1 protein in the protoplasts of Arabidopsis A:The green fluorescent signals of the HaLACS1 gene. B:Endoplasmic reticulum marker. C:Bright field. D:Merged
| [1] | 马惠茹, 等. 内蒙古河套地区向日葵饲料资源生产情况及开发利用现状[J]. 中国畜牧兽医, 2014, 41(3):251-254. |
| Ma HR, et al. Development and production of sunflower by-product feed resource in Hetao area in Inner Mongolia[J]. China Animal Husb Vet Med, 2014, 41(3):251-254. | |
| [2] | 汪磊, 谭美莲, 傅春玲, 等. 利用近红外技术预测向日葵籽仁品质性状[J]. 中国油料作物学报, 2020, 42(1):147-153. |
| Wang L, Tan ML, Fu CL, et al. Prediction of qualitative characteristics of sunflower husked seed by near infrared spectroscopy[J]. Chin J Oil Crop Sci, 2020, 42(1):147-153. | |
| [3] | Hryvusevich P, Navaselsky I, Talkachova Y, et al. Sodium influx and potassium efflux currents in sunflower root cells under high salinity[J]. Front Plant Sci, 2020, 11:613936. |
| [4] |
Kelly AA, Quettier AL, Shaw E, et al. Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis[J]. Plant Physiol, 2011, 157(2):866-875.
doi: 10.1104/pp.111.181784 URL |
| [5] |
Ding LN, Gu SL, Zhu FG, et al. Long-chain acyl-CoA synthetase 2 is involved in seed oil production in Brassica napus[J]. BMC Plant Biol, 2020, 20(1):21.
doi: 10.1186/s12870-020-2240-x URL |
| [6] |
Groot PH, et al. Fatty acid activation:specificity, localization, and function[J]. Adv Lipid Res, 1976, 14:75-126.
pmid: 3952 |
| [7] |
Hayashi H, De Bellis L, Hayashi Y, et al. Molecular characterization of an Arabidopsis acyl-coenzyme a synthetase localized on glyoxysomal membranes[J]. Plant Physiol, 2002, 130(4):2019-2026.
doi: 10.1104/pp.012955 URL |
| [8] |
Pei ZT, Oey NA, Zuidervaart MM, et al. The acyl-CoA synthetase “bubblegum”(lipidosin):Further characterization and role in neuronal fatty acid β-oxidation[J]. J Biol Chem, 2003, 278(47):47070-47078.
doi: 10.1074/jbc.M310075200 URL |
| [9] |
Babbitt PC, Kenyon GL, Martin BM, et al. Ancestry of the 4-chlorobenzoate dehalogenase:analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases[J]. Biochemistry, 1992, 31(24):5594-5604.
pmid: 1351742 |
| [10] |
Iijima H, Fujino T, Minekura H, et al. Biochemical studies of two rat acyl-CoA synthetases, ACS1 and ACS2[J]. Eur J Biochem, 1996, 242(2):186-190.
pmid: 8973631 |
| [11] |
Steinberg SJ, Morgenthaler J, Heinzer AK, et al. Very long-chain acyl-CoA synthetases:human “bubblegum” represents a new family of proteins capable of activating very long-chain fatty acids[J]. J Biol Chem, 2000, 275(45):35162-35169.
doi: 10.1074/jbc.M006403200 pmid: 10954726 |
| [12] |
Shockey JM, Fulda MS, Browse JA. Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism[J]. Plant Physiol, 2002, 129(4):1710-1722.
pmid: 12177484 |
| [13] |
Zhao L, Katavic V, Li F, et al. Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1(LACS1), but not LACS8, functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis[J]. Plant J, 2010, 64(6):1048-1058.
doi: 10.1111/j.1365-313X.2010.04396.x URL |
| [14] |
Lv SY, Song T, Kosma DK, et al. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1(LACS1)that has overlapping functions with LACS2 in plant wax and cutin synthesis[J]. Plant J, 2009, 59(4):553-564.
doi: 10.1111/j.1365-313X.2009.03892.x URL |
| [15] | Zhao HY, Kosma DK, Lü S. Functional role of long-chain acyl-CoA synthetases in plant development and stress responses[J]. Front Plant Sci, 2021, 12:640996. |
| [16] |
Weng H, Molina I, Shockey J, et al. Organ fusion and defective cuticle function in a Lacs1 Lacs2 double mutant of Arabidopsis[J]. Planta, 2010, 231(5):1089-1100.
doi: 10.1007/s00425-010-1110-4 pmid: 20237894 |
| [17] |
Zhao LF, Haslam TM, Sonntag A, et al. Functional overlap of long-chain acyl-CoA synthetases in Arabidopsis[J]. Plant Cell Physiol, 2019, 60(5):1041-1054.
doi: 10.1093/pcp/pcz019 URL |
| [18] |
Faergeman NJ, Black PN, Zhao XD, et al. The Acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular Utilization[J]. J Biol Chem, 2001, 276(40):37051-37059.
doi: 10.1074/jbc.M100884200 pmid: 11477098 |
| [19] |
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR[J]. Nucleic Acids Res, 2001, 29(9):e45.
doi: 10.1093/nar/29.9.e45 pmid: 11328886 |
| [20] | Mereshchuk A, Chew JSK, Dobson MJ. Use of yeast plasmids:transformation and inheritance assays[J]. Methods Mol Biol, 2021, 2196:1-13. |
| [21] |
Bua A, et al. Antimicrobial activity of Austroeupatorium inulaefolium(H. B. K. )against intracellular and extracellular organisms[J]. Nat Prod Res, 2018, 32(23):2869-2871.
doi: 10.1080/14786419.2017.1385014 pmid: 29017356 |
| [22] | Donadu M, Usai D, et al. In vitro activity of hybrid lavender essential oils against multidrug resistant strains of Pseudomonas aeruginosa[J]. J Infect Dev Countr, 2018, 12(1):9-14. |
| [23] |
宋燕子, 贾彬, 等. 莱茵衣藻酰基辅酶A合成酶cDNA克隆及其酵母表达[J]. 生物技术通报, 2015, 31(9):119-124.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.09.016 |
| Song YZ, Jia B, et al. cDNA cloning and yeast expression of acyl-CoA synthetase of Chlamydomonas reinhardtii[J]. Biotechnol Bull, 2015, 31(9):119-124. | |
| [24] | 李艾. 斯氏油脂酵母细胞内油脂含量的快速检测方法研究[J]. 中国油脂, 2016, 41(11):79-82. |
| Li A. Rapid determination of oil content in Lipomyces Starkeyi cells[J]. China Oils Fats, 2016, 41(11):79-82. | |
| [25] |
Wang XL, Li XB. The GhACS1 gene encodes an acyl-CoA synthetase which is essential for normal microsporogenesis in early anther development of cotton[J]. Plant J, 2009, 57(3):473-486.
doi: 10.1111/j.1365-313X.2008.03700.x URL |
| [26] |
Jessen D, Roth C, Wiermer M, et al. Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis[J]. Plant Physiol, 2015, 167(2):351-366.
doi: 10.1104/pp.114.250365 pmid: 25540329 |
| [27] |
Schnurr JA, Shockey JM, de Boer GJ, et al. Fatty acid export from the chloroplast. molecular characterization of a major plastidial acyl-coenzyme a synthetase from Arabidopsis[J]. Plant Physiol, 2002, 129(4):1700-1709.
pmid: 12177483 |
| [28] | Yu L, Tan X, Jiang B, et al. A peroxisomal long-chain acyl-CoA synthetase from Glycine max involved in lipid degradation[J]. PLoS One, 2014, 9(7):e100144. |
| [29] |
Aznar-Moreno JA, Venegas Calerón M, Martínez-Force E, et al. Sunflower(Helianthus annuus)long-chain acyl-coenzyme A synthetases expressed at high levels in developing seeds[J]. Physiol Plant, 2014, 150(3):363-373.
doi: 10.1111/ppl.12107 pmid: 24102504 |
| [30] |
Jessen D, Olbrich A, Knüfer J, et al. Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis[J]. Plant J, 2011, 68(4):715-726.
doi: 10.1111/j.1365-313X.2011.04722.x URL |
| [31] |
Pulsifer IP, Kluge S, Rowland O. Arabidopsis long-chain acyl-CoA synthetase 1(LACS1), LACS2, and LACS3 facilitate fatty acid uptake in yeast[J]. Plant Physiol Biochem, 2012, 51:31-39.
doi: 10.1016/j.plaphy.2011.10.003 URL |
| [32] |
McFarlane HE, Watanabe Y, et al. Golgi- and trans-Golgi network-mediated vesicle trafficking is required for wax secretion from epidermal cells[J]. Plant Physiol, 2014, 164(3):1250-1260.
doi: 10.1104/pp.113.234583 pmid: 24468625 |
| [33] |
Zhang CL, Mao K, Zhou LJ, et al. Genome-wide identification and characterization of apple long-chain Acyl-CoA synthetases and expression analysis under different stresses[J]. Plant Physiol Biochem, 2018, 132:320-332.
doi: 10.1016/j.plaphy.2018.09.004 URL |
| [34] | Zhang CL, et al. An apple long-chain acyl-CoA synthetase, MdLACS4, induces early flowering and enhances abiotic stress resistance in Arabidopsis[J]. Plant Sci, 2020, 297:110529. |
| [35] |
Cominelli E, Sala TA, Calvi D, et al. Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability[J]. Plant J, 2008, 53(1):53-64.
pmid: 17971045 |
| [1] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [2] | SONG Zhi-zhong, XU Wei-hua, XIAO Hui-lin, TANG Mei-ling, CHEN Jing-hui, GUAN Xue-qiang, LIU Wan-hao. Cloning, Expression and Function of Iron Regulated Transporter VvIRT1 in Wine Grape(Vitis vinifera L.) [J]. Biotechnology Bulletin, 2023, 39(8): 234-240. |
| [3] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [4] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [5] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [6] | HAO Qing-qing, YAO Sheng, LIU Jia-he, CHEN Pei-zhen, ZHANG Meng-yang, JI Kong-shu. Cloning and Expression Analysis of NAC Transcription Factor PmNAC8 in Pinus massoniana [J]. Biotechnology Bulletin, 2022, 38(4): 202-216. |
| [7] | DANG Yuan, LI Wei, MIAO Xiang, XIU Yu, LIN Shan-zhi. Cloning of Oleosin Gene PsOLE4 from Prunus sibirica and Its Regulatory Function Analysis for Oil Accumulation [J]. Biotechnology Bulletin, 2022, 38(11): 151-161. |
| [8] | LUO Ying, TANG Zhi, WANG Fan, LIU Xiao-xia, LUO Xiao-fang, HE Fu-lin. Cloning and Response Analysis to Abiotic Stress of GbR2R3-MYB1 Gene from Ginkgo biloba [J]. Biotechnology Bulletin, 2022, 38(10): 184-194. |
| [9] | SUN Rui-fen, ZHANG Yan-fang, NIU Su-qing, GUO Shu-chun, LI Su-ping, YU Hai-feng, NIE Hui, MOU Ying-nan. Expression Analysis and Functional Verification of the HaACO1 Gene in Sunflower [J]. Biotechnology Bulletin, 2021, 37(9): 114-124. |
| [10] | FAN Ya-peng, RUI Cun, ZHANG Yue-xin, CHEN Xiu-gui, LU Xu-ke, WANG Shuai, ZHANG Hong, XU Nan, WANG Jing, CHEN Chao, YE Wu-wei. Cloning,Expression and Preliminary Bioinformatics Analysis of the Alkaline Tolerant Gene GhZAT12 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(8): 121-130. |
| [11] | SHAN Cao-mei, YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang. Cloning,and Functional Analysis of Gene OsRAI1 Resistant to Hirschmanniella mucronate in Rice [J]. Biotechnology Bulletin, 2021, 37(7): 146-155. |
| [12] | SUN Xiao-qian, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Gene Cloning,Subcellular Localization and Expression Analyses of FtMYBF Transcription Factor in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2021, 37(3): 10-17. |
| [13] | HAN Zhan-hong, ZONG Yuan-yuan, ZHANG Xue-mei, WANG Bin, PRUSKY Dov, BI Yang. Bioinformatics,Subcellular Localization and Expression Analysis of erg4 in Penicillium expansum [J]. Biotechnology Bulletin, 2021, 37(12): 60-70. |
| [14] | ZHANG Rui-zhu, JIANG Yu-chen, HUANG Jun, YAN Jie. Cloning and Expression Analysis of SRPP2 Gene in Taraxacum kok-saghyz Rodin [J]. Biotechnology Bulletin, 2020, 36(1): 9-14. |
| [15] | DUAN Min-jie, YI Hong-wei, WANG Jin, WU Zheng. Cloning and Expression Analysis of the Pear Black Spot Resistant Gene PpEMS1 [J]. Biotechnology Bulletin, 2019, 35(11): 16-21. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||