








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (7): 62-69.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1592
Previous Articles Next Articles
SHI Jia( ), ZHU Xiu-mei, XUE Meng-yu, YU Chao, WEI Yi-ming, YANG Feng-huan, CHEN Hua-min(
), ZHU Xiu-mei, XUE Meng-yu, YU Chao, WEI Yi-ming, YANG Feng-huan, CHEN Hua-min( )
)
Received:2021-12-25
Online:2022-07-26
Published:2022-08-09
Contact:
CHEN Hua-min
E-mail:qwer1768004860@163.com;chenhuamin@caas.cn
SHI Jia, ZHU Xiu-mei, XUE Meng-yu, YU Chao, WEI Yi-ming, YANG Feng-huan, CHEN Hua-min. Optimization and Application of the Chromatin Immunoprecipitation Based on Rice Protoplast[J]. Biotechnology Bulletin, 2022, 38(7): 62-69.
| 家族Family | 基因Gene | ID | CCAAT 数目及位置CCAAT number and position |
|---|---|---|---|
| OsNRT1家族(低亲和力)OsNRT1 family(low affinity) | OsNRT1.1 | Os03g13274 | 2(-1404,-1055) |
| OsNRT2家族(高亲和力)OsNRT2 family(high affinity) | OsNRT2.1 | Os02g02170 | 1(-175) |
| OsNRT2.2 | Os02g02190 | 2(-276,-53) | |
| OsNAR2.1 | Os02g38230 | 2(-631,-460) |
Table 1 Prediction of regulatory elements in the promoter region of NRTs genes
| 家族Family | 基因Gene | ID | CCAAT 数目及位置CCAAT number and position |
|---|---|---|---|
| OsNRT1家族(低亲和力)OsNRT1 family(low affinity) | OsNRT1.1 | Os03g13274 | 2(-1404,-1055) |
| OsNRT2家族(高亲和力)OsNRT2 family(high affinity) | OsNRT2.1 | Os02g02170 | 1(-175) |
| OsNRT2.2 | Os02g02190 | 2(-276,-53) | |
| OsNAR2.1 | Os02g38230 | 2(-631,-460) |
| 基因名称 Gene name | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 片段大小及内容 Segment size and content |
|---|---|---|---|
| OsNRT1.1 | OsNRT1.1 Q-F2 | TGCTACGGTCTCATCTTCTCT | 199 bp 包含NRT1.1基因启动子序列中的第二个CCAAT-box 199 bp contains the second CCAAT-box in the promoter of NRT1.1 |
| OsNRT1.1 Q-R2 | CAAGTAATCCATCTAACGCTACGA | ||
| OsNRT2.1 | OsNRT2.1 Q-F1 | ACGAATCTTAAGGCAAAT | 194 bp包含NRT2.1基因启动子序列中的一个CCAAT-box 199 bp contains a CCAAT-box in the promoter of NRT2.1 |
| OsNRT2.1 Q-R1 | CTTCTTGGAGATGGAATC | ||
| OsNRT2.2 | OsNRT2.2 Q-F2 | CACGAGGCAGATATTACAACTTGA | 149 bp 包含NRT2.2基因启动子序列中的第二个CCAAT-box 149 bp contains the second CCAAT-box in the promoter of NRT2.2 |
| OsNRT2.2 Q-R2 | CGGTGACGATGATCTTGGC | ||
| OsNAR2.1 | OsNAR2.1 Q-F2 | TTCCTCCATTAAGAACGCCTTC | 119 bp 包含NAR2.1基因启动子序列中的第二个CCAAT-box 119 bp contains the second CCAAT-box in the promoter of NAR2.1 |
| OsNAR2.1 Q-R2 | TTGAGTGCCTCGGTTGTTG | ||
| Negative | OsNAR2.1-NF | GATGGCTGTCCTGCTCTTG | 183 bp 为NAR2.1基因启动子中不含CCAAT-box的序列 183 bp NAR2.1 gene promoter without CCAAT-box |
| OsNAR2.1-NR | ATCATTTCGCTCCTCCAAACTATT |
Table 2 Required primer sequences for RT-qPCR validation
| 基因名称 Gene name | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 片段大小及内容 Segment size and content |
|---|---|---|---|
| OsNRT1.1 | OsNRT1.1 Q-F2 | TGCTACGGTCTCATCTTCTCT | 199 bp 包含NRT1.1基因启动子序列中的第二个CCAAT-box 199 bp contains the second CCAAT-box in the promoter of NRT1.1 |
| OsNRT1.1 Q-R2 | CAAGTAATCCATCTAACGCTACGA | ||
| OsNRT2.1 | OsNRT2.1 Q-F1 | ACGAATCTTAAGGCAAAT | 194 bp包含NRT2.1基因启动子序列中的一个CCAAT-box 199 bp contains a CCAAT-box in the promoter of NRT2.1 |
| OsNRT2.1 Q-R1 | CTTCTTGGAGATGGAATC | ||
| OsNRT2.2 | OsNRT2.2 Q-F2 | CACGAGGCAGATATTACAACTTGA | 149 bp 包含NRT2.2基因启动子序列中的第二个CCAAT-box 149 bp contains the second CCAAT-box in the promoter of NRT2.2 |
| OsNRT2.2 Q-R2 | CGGTGACGATGATCTTGGC | ||
| OsNAR2.1 | OsNAR2.1 Q-F2 | TTCCTCCATTAAGAACGCCTTC | 119 bp 包含NAR2.1基因启动子序列中的第二个CCAAT-box 119 bp contains the second CCAAT-box in the promoter of NAR2.1 |
| OsNAR2.1 Q-R2 | TTGAGTGCCTCGGTTGTTG | ||
| Negative | OsNAR2.1-NF | GATGGCTGTCCTGCTCTTG | 183 bp 为NAR2.1基因启动子中不含CCAAT-box的序列 183 bp NAR2.1 gene promoter without CCAAT-box |
| OsNAR2.1-NR | ATCATTTCGCTCCTCCAAACTATT |
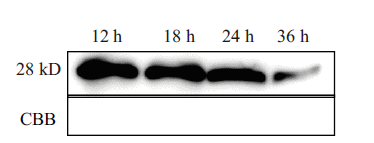
Fig.2 Expressions detection of OsNF-YA4 protein after protoplast transformed After rice protoplast transformed with expression vector and the incubation was performed at different times,the transient expressed proteins were extracted,and the expression of fused protein was detected via anti-Myc antibody 1-4:NF-YA4 protein expressions at 12,18,24 and 36 h. Coomassie brilliant blue staining showed the loaded protein in each lane
| 甲醛终浓度 Final concentration of formaldehyde/% | 细胞活性百分比 Percentage of cell activity/% |
|---|---|
| 0 | 100+0.01 |
| 0.1 | 93+2.79 |
| 0.3 | 80+4.27 |
| 0.7 | 72+3.98 |
| 1.0 | 48+2.72 |
Table 3 Effects of different final concentrations of formal-dehyde on protoplasmic cell activity
| 甲醛终浓度 Final concentration of formaldehyde/% | 细胞活性百分比 Percentage of cell activity/% |
|---|---|
| 0 | 100+0.01 |
| 0.1 | 93+2.79 |
| 0.3 | 80+4.27 |
| 0.7 | 72+3.98 |
| 1.0 | 48+2.72 |
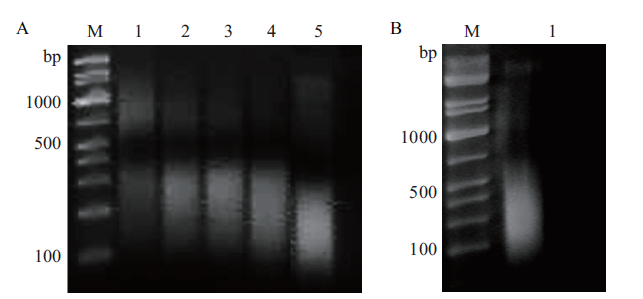
Fig.3 Optimization of ultrasonic fragmentation time on OsNF-YA4 ChIPs-enriched DNA(A)and validation of the fragmentation(B) A:Effects of ultrasonic crushing time on fragment length(M:DL2000 marker;1-5:ultrasonic duration was 5,10,15,20 and 30 min respectivey)B:The fragmentation of DNA enrichment by ChIPs(1:the fragmentation of input when ultrasonic treated 15 min)
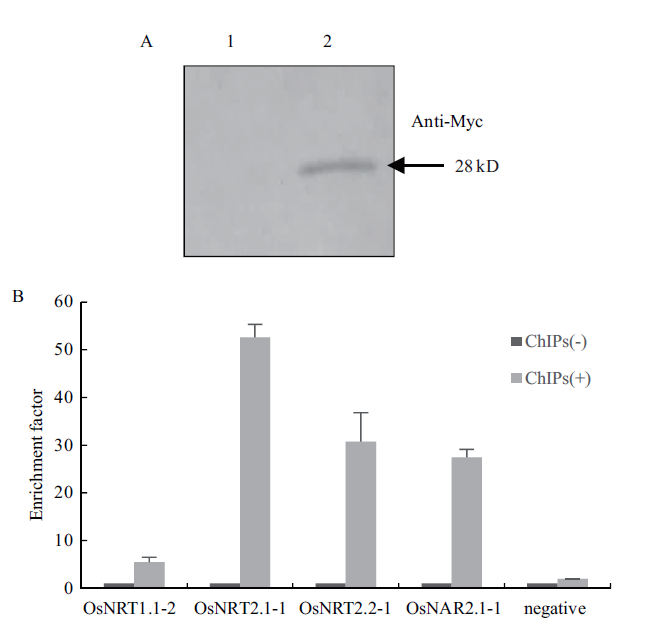
Fig.4 Detection of the effect of chromosome immunopreci-pitation(A)and specificity analysis of enriched DNA(B)A:The level of OsNFYA4 after immunoprecipitation detected by Western blot(1:immunoprecipitated samples(ChIPs-)without anti-Myc antibody;2:immunoprecipitated samples with anti-Myc antibody(ChIPs+)). B:Validation of the specificity of the ChIP-RP enriched DNA by qRT-PCR
| [1] |
Mitsuda N, Ohme-Takagi M. Functional analysis of transcription factors in Arabidopsis[J]. Plant Cell Physiol, 2009, 50(7):1232-1248.
doi: 10.1093/pcp/pcp075 URL |
| [2] |
Zou YM, Wang SF, Zhou YY, et al. Transcriptional regulation of the immune receptor FLS2 controls the ontogeny of plant innate immunity[J]. Plant Cell, 2018, 30(11):2779-2794.
doi: 10.1105/tpc.18.00297 URL |
| [3] |
Bannister AJ, Kouzarides T. Basic peptides enhance protein/DNA interaction in vitro[J]. Nucleic Acids Res, 1992, 20(13):3523.
pmid: 1630932 |
| [4] |
Wang MM, Reed RR. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast[J]. Nature, 1993, 364(6433):121-126.
doi: 10.1038/364121a0 URL |
| [5] |
杨立文, 刘双荣, 李玉红, 等. 植物转录因子与DNA互作研究技术[J]. 植物学报, 2020, 55(4):468-474.
doi: 10.11983/CBB20057 |
| Yang LW, Liu SR, Li YH, et al. Methods for examining transcription factor-DNA interaction in plants[J]. Chin Bull Bot, 2020, 55(4):468-474. | |
| [6] |
Gilmour DS, Lis JT. Detecting protein-DNA interactions in vivo:distribution of RNA polymerase on specific bacterial genes[J]. Proc Natl Acad Sci USA, 1984, 81(14):4275-4279.
doi: 10.1073/pnas.81.14.4275 URL |
| [7] |
Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking:a probe for in vivo chromatin structures[J]. Proc Natl Acad Sci USA, 1985, 82(19):6470-6474.
doi: 10.1073/pnas.82.19.6470 URL |
| [8] |
Mundade R, Ozer HG, Wei H, et al. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond[J]. Cell Cycle, 2014, 13(18):2847-2852.
doi: 10.4161/15384101.2014.949201 pmid: 25486472 |
| [9] |
Kaufmann K, Muiño JM, Østerås M, et al. Chromatin immunoprecipitation(ChIP)of plant transcription factors followed by sequencing(ChIP-SEQ)or hybridization to whole genome arrays(ChIP-CHIP)[J]. Nat Protoc, 2010, 5(3):457-472.
doi: 10.1038/nprot.2009.244 pmid: 20203663 |
| [10] |
Nardini M, Gnesutta N, Donati G, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination[J]. Cell, 2013, 152(1/2):132-143.
doi: 10.1016/j.cell.2012.11.047 URL |
| [11] |
Zhao BT, Ge LF, Liang RQ, et al. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor[J]. BMC Mol Biol, 2009, 10:29.
doi: 10.1186/1471-2199-10-29 URL |
| [12] |
Luan MD, Xu MY, Lu YM, et al. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves[J]. Gene, 2015, 555(2):178-185.
doi: 10.1016/j.gene.2014.11.001 URL |
| [13] |
Ding Q, Zeng J, He XQ. MiR169 and its target PagHAP2-6 regulated by ABA are involved in poplar cambium dormancy[J]. J Plant Physiol, 2016, 198:1-9.
doi: 10.1016/j.jplph.2016.03.017 URL |
| [14] |
Li WX, Oono Y, Zhu JH, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance[J]. Plant Cell, 2008, 20(8):2238-2251.
doi: 10.1105/tpc.108.059444 URL |
| [15] |
Zhao M, Ding H, Zhu JK, et al. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis[J]. New Phytol, 2011, 190(4):906-915.
doi: 10.1111/j.1469-8137.2011.03647.x URL |
| [16] |
Combier JP, Frugier F, de Billy F, et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula[J]. Genes Dev, 2006, 20(22):3084-3088.
doi: 10.1101/gad.402806 URL |
| [17] |
Warpeha KM, Upadhyay S, Yeh J, et al. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis[J]. Plant Physiol, 2007, 143(4):1590-1600.
doi: 10.1104/pp.106.089904 URL |
| [18] |
Xu MY, Zhang L, Li WW, et al. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana[J]. J Exp Bot, 2013, 65(1):89-101.
doi: 10.1093/jxb/ert353 URL |
| [19] | Para A, Li Y, Coruzzi GM. μChIP-seq for genome-wide mapping of in vivo TF-DNA interactions in Arabidopsis root protoplasts[J]. Methods Mol Biol Clifton N J, 2018, 1761:249-261. |
| [20] | He F, Zhang F, Sun WX, et al. A versatile vector toolkit for functional analysis of rice genes[J]. Rice(N Y), 2018, 11(1):27. |
| [21] | Chen HM, Zou Y, Shang YL, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants[J]. Plant Physiol, 2008, 146(2):368-376. |
| [22] |
Yu C, Chen YT, Cao YQ, et al. Overexpression of miR169o, an overlapping microRNA in response to both nitrogen limitation and bacterial infection, promotes nitrogen use efficiency and susceptibility to bacterial blight in rice[J]. Plant Cell Physiol, 2018, 59(6):1234-1247.
doi: 10.1093/pcp/pcy060 URL |
| [23] | 翁小煜, 周少立, 宗伟, 等. 水稻染色质免疫共沉淀[M]. Bio-protocol, 2018, Bio-101:e1010135. |
|
Weng XY, Zhou SL, Zong W, et al. ChIP assay in rice[M]. Bio-protocol, 2018, Bio-101:e1010135. DOI: 10.21769/BioProtoc.1010135.
doi: 10.21769/BioProtoc.1010135 |
|
| [24] |
Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1):325-327.
doi: 10.1093/nar/30.1.325 URL |
| [25] |
Baranello L, Kouzine F, Sanford S, et al. ChIP bias as a function of cross-linking time[J]. Chromosome Res, 2016, 24(2):175-181.
doi: 10.1007/s10577-015-9509-1 pmid: 26685864 |
| [26] |
Ricardi MM, González RM, Iusem ND. Protocol:fine-tuning of a Chromatin Immunoprecipitation(ChIP)protocol in tomato[J]. Plant Methods, 2010, 6:11.
doi: 10.1186/1746-4811-6-11 URL |
| [27] | 董浩, 彭小薇, 王晓英, 等. 染色质免疫共沉淀技术对羊种布鲁氏菌转录调控因子MucR靶基因的筛选[J]. 中国农业大学学报, 2016, 21(4):102-106. |
| Dong H, Peng XW, Wang XY, et al. Identification of the targets of MucR by chromatin immunoprecipitation in Brucella melitensis[J]. J China Agric Univ, 2016, 21(4):102-106. |
| [1] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [2] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [3] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [4] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [5] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [6] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [7] | GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L. [J]. Biotechnology Bulletin, 2023, 39(6): 217-232. |
| [8] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [9] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [10] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| [11] | GE Yan-rui, ZHAO Ran, XU Jing, LI Ruo-fan, HU Yun-tao, LI Rui-li. Advances in the Development and Regulation of Vascular Cambium [J]. Biotechnology Bulletin, 2023, 39(3): 13-25. |
| [12] | LIU Cheng-xia, SUN Zong-yan, LUO Yun-bo, ZHU Hong-liang, QU Gui-qin. Multifaceted Roles of bHLH Phosphorylation in Regulation of Plant Physiological Functions [J]. Biotechnology Bulletin, 2023, 39(3): 26-34. |
| [13] | ZHAO Meng-liang, GUO Yi-ting, REN Yan-jing. Identification and Analysis of WRKY Transcription Factor Family Genes in Helianthus tuberosus [J]. Biotechnology Bulletin, 2023, 39(2): 116-125. |
| [14] | HAN Fang-ying, HU Xin, WANG Nan-nan, XIE Yu-hong, WANG Xiao-yan, ZHU Qiang. Research Progress in Response of DREBs to Abiotic Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(11): 86-98. |
| [15] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||