








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (7): 51-61.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1524
Previous Articles Next Articles
CHEN Jia-min1,2( ), LIU Yong-jie2, MA Jin-xiu2, LI Dan2, GONG Jie2, ZHAO Chang-ping2, GENG Hong-wei1(
), LIU Yong-jie2, MA Jin-xiu2, LI Dan2, GONG Jie2, ZHAO Chang-ping2, GENG Hong-wei1( ), GAO Shi-qing2(
), GAO Shi-qing2( )
)
Received:2021-12-08
Online:2022-07-26
Published:2022-08-09
Contact:
GENG Hong-wei,GAO Shi-qing
E-mail:cjmin2018@126.com;hw-geng@163.com;gshiq@126.com
CHEN Jia-min, LIU Yong-jie, MA Jin-xiu, LI Dan, GONG Jie, ZHAO Chang-ping, GENG Hong-wei, GAO Shi-qing. Expression Pattern Analysis of Histone Methyltransferase Under Drought Stress in Hybrid Wheat[J]. Biotechnology Bulletin, 2022, 38(7): 51-61.
| 引物名称Primer name | 上游引物序列 Forward primer sequence(5'-3') | 下游引物序列 Reverse primer sequence(5'-3') |
|---|---|---|
| actin | GTTGGTGATGAGGCCCAATC | GTGCTACACGGAGCTCATTG |
| TaHMT21 | TGCCGCTGTGGTGTATACTG | CAGCCAAAAGACCCCATCCA |
| TaHMT24 | TAAGGCTAGGGGCAACTCCT | CCATCCAACTCAGCAGTCGT |
| TaHMT31 | GTTTGGTGGTGTGATGGTGC | TGCTGGAATGGTGTCCTTCG |
| TaHMT42 | GGTCTGCGCAAGCAATTGAA | TCGTACAGGGCGTCGATTTC |
| TaHMT49 | TTCTTCACGAGCAGGAAGGT | TTCCTCGGCAGTACTTGCTT |
| TaHMT105 | AGACCTTAGAGAGCGGCGTA | ATGCTCATCCCCGAGAACAC |
| TaHMT143 | TGATTGCAGCATGGACCAGT | GCAAACAGCCCCAAAGTACG |
| TaHMT157 | CTACGGTTGCATCAAGCTCA | GCATCTCACAGTCCTCGTCA |
Table 1 Primer sequences for real-time quantitative PCR
| 引物名称Primer name | 上游引物序列 Forward primer sequence(5'-3') | 下游引物序列 Reverse primer sequence(5'-3') |
|---|---|---|
| actin | GTTGGTGATGAGGCCCAATC | GTGCTACACGGAGCTCATTG |
| TaHMT21 | TGCCGCTGTGGTGTATACTG | CAGCCAAAAGACCCCATCCA |
| TaHMT24 | TAAGGCTAGGGGCAACTCCT | CCATCCAACTCAGCAGTCGT |
| TaHMT31 | GTTTGGTGGTGTGATGGTGC | TGCTGGAATGGTGTCCTTCG |
| TaHMT42 | GGTCTGCGCAAGCAATTGAA | TCGTACAGGGCGTCGATTTC |
| TaHMT49 | TTCTTCACGAGCAGGAAGGT | TTCCTCGGCAGTACTTGCTT |
| TaHMT105 | AGACCTTAGAGAGCGGCGTA | ATGCTCATCCCCGAGAACAC |
| TaHMT143 | TGATTGCAGCATGGACCAGT | GCAAACAGCCCCAAAGTACG |
| TaHMT157 | CTACGGTTGCATCAAGCTCA | GCATCTCACAGTCCTCGTCA |

Fig.1 Heatmap of TaHMT candidate gene expressions under drought stress in wheat A:Venn diagram of differentially expressed genes and homologous genes on Wheat Expression Brower database and WheatOmics database. B:Expression heatmap of 8 candidate genes in Wheat Expression Brower database. C:Expression heatmap of 8 candidate genes in WheatOmics database. The red font is the candidate gene
| 基因名称Gene name | TaHMT21 | TaHMT24 | TaHMT31 | TaHMT42 | TaHMT49 | TaHMT105 | TaHMT143 | TaHMT157 |
|---|---|---|---|---|---|---|---|---|
| 基因ID号Gene ID | TraesCS2A02G302100 | TraesCS2A02G457100 | TraesCS2B02G317900 | TraesCS2D02G300800 | TraesCS2D02G566400 | TraesCS5B02G163000 | TraesCS7B02G262900 | TraesCS2A02G147100 |
| 染色体位置Chromosome location | 2A:517 803 993-517 808 473(+) | 2A:705 740 079-705 742 875(+) | 2B:453 747 757-453 752 627(+) | 2D:383 285 805-383 290 377(+) | 2D:635 593 374-635 597 307(-) | 5B:300 380 740-300 397 790(-) | 7B:483 766 747-483 769 174(-) | 2A:93 926 568-93 930 236(-) |
| 亚细胞定位Subcellular localization | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus |
| 氨基酸数目Number of amino acids/aa | 502 | 476 | 529 | 502 | 737 | 1086 | 390 | 601 |
| 分子量Molecular weight/kD | 56 612.99 | 53 938.51 | 59 323.00 | 56 560.96 | 80 411.70 | 122 007.19 | 42 293.23 | 65 280.55 |
| 理论等电点Theoretical pI | 4.77 | 5.47 | 4.78 | 4.80 | 7.09 | 7.59 | 4.44 | 4.48 |
| 负电荷残基数(Asp+Glu) Number of negatively charged residues(Asp+Glu) | 83 | 64 | 84 | 82 | 84 | 146 | 65 | 99 |
| 正电荷残基数(Arg+Lys) Number of positively charged residues(Arg+Lys) | 53 | 53 | 53 | 53 | 83 | 148 | 30 | 50 |
| 总原子数Total number of atoms | 7 801 | 7 540 | 8 162 | 7 794 | 11 137 | ------ | 5 789 | 8 992 |
| 分子式Formula | C2458H3830N684O 805S24 | C2392H3751N657O 716S24 | C2572H4003N717O 843S27 | C2458H3826N684O 802S24 | C3478H5503N1035O 1080S41 | ------ | C1833H2830N506O 594S26 | C2859H4408N762O 937S26 |
| 不稳定指数Instability index | 51.39 | 43.26 | 49.28 | 53.18 | 49.44 | 54.44 | 54.59 | 38.91 |
| 脂溶指数Aliphatic index | 71.14 | 90.82 | 71.02 | 71.53 | 68.41 | 76.19 | 75.95 | 75.61 |
| 亲水性GRAVY | -0.631 | -0.219 | -0.584 | -0.608 | -0.511 | -0.514 | -0.301 | -0.285 |
Table 2 Basic information of TaHMT drought stress candidate genes
| 基因名称Gene name | TaHMT21 | TaHMT24 | TaHMT31 | TaHMT42 | TaHMT49 | TaHMT105 | TaHMT143 | TaHMT157 |
|---|---|---|---|---|---|---|---|---|
| 基因ID号Gene ID | TraesCS2A02G302100 | TraesCS2A02G457100 | TraesCS2B02G317900 | TraesCS2D02G300800 | TraesCS2D02G566400 | TraesCS5B02G163000 | TraesCS7B02G262900 | TraesCS2A02G147100 |
| 染色体位置Chromosome location | 2A:517 803 993-517 808 473(+) | 2A:705 740 079-705 742 875(+) | 2B:453 747 757-453 752 627(+) | 2D:383 285 805-383 290 377(+) | 2D:635 593 374-635 597 307(-) | 5B:300 380 740-300 397 790(-) | 7B:483 766 747-483 769 174(-) | 2A:93 926 568-93 930 236(-) |
| 亚细胞定位Subcellular localization | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus |
| 氨基酸数目Number of amino acids/aa | 502 | 476 | 529 | 502 | 737 | 1086 | 390 | 601 |
| 分子量Molecular weight/kD | 56 612.99 | 53 938.51 | 59 323.00 | 56 560.96 | 80 411.70 | 122 007.19 | 42 293.23 | 65 280.55 |
| 理论等电点Theoretical pI | 4.77 | 5.47 | 4.78 | 4.80 | 7.09 | 7.59 | 4.44 | 4.48 |
| 负电荷残基数(Asp+Glu) Number of negatively charged residues(Asp+Glu) | 83 | 64 | 84 | 82 | 84 | 146 | 65 | 99 |
| 正电荷残基数(Arg+Lys) Number of positively charged residues(Arg+Lys) | 53 | 53 | 53 | 53 | 83 | 148 | 30 | 50 |
| 总原子数Total number of atoms | 7 801 | 7 540 | 8 162 | 7 794 | 11 137 | ------ | 5 789 | 8 992 |
| 分子式Formula | C2458H3830N684O 805S24 | C2392H3751N657O 716S24 | C2572H4003N717O 843S27 | C2458H3826N684O 802S24 | C3478H5503N1035O 1080S41 | ------ | C1833H2830N506O 594S26 | C2859H4408N762O 937S26 |
| 不稳定指数Instability index | 51.39 | 43.26 | 49.28 | 53.18 | 49.44 | 54.44 | 54.59 | 38.91 |
| 脂溶指数Aliphatic index | 71.14 | 90.82 | 71.02 | 71.53 | 68.41 | 76.19 | 75.95 | 75.61 |
| 亲水性GRAVY | -0.631 | -0.219 | -0.584 | -0.608 | -0.511 | -0.514 | -0.301 | -0.285 |
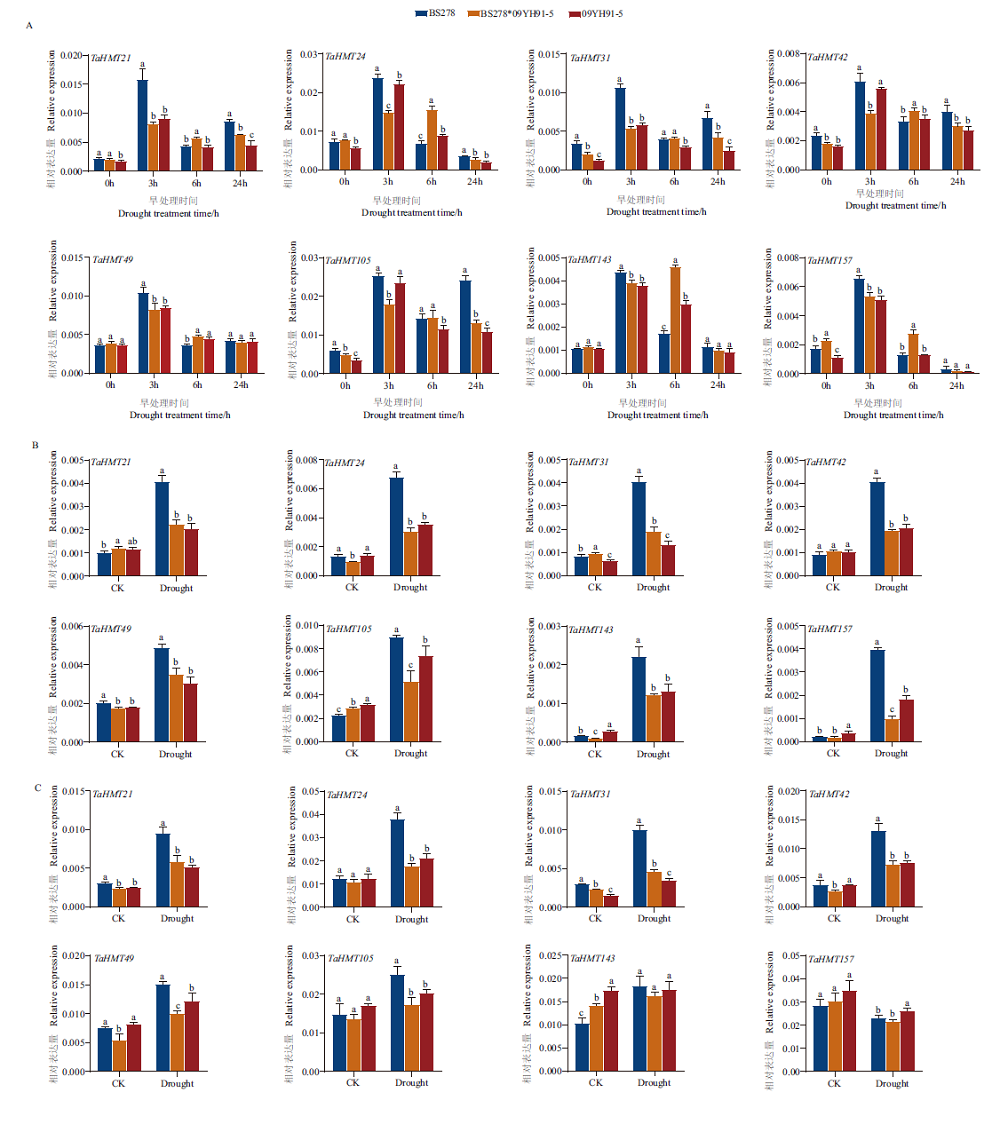
Fig.6 Expressions of TaHMT under drought stress A:Gene expression in the wheat leaves during vegetative growth. B:Gene expression in the shoot of wheat at seedling stage. C:Gene expression in the underground part of wheat at seedling stage. The error line in the figure refers to the standard deviation. The lowercase letters indicate significant differences between different materials at the same time(P<0.05)
| [1] | Goldberg AD, Allis CD, Bernstein E. Epigenetics:a landscape takes shape[J]. Cell, 2007, 128(4):635-638. |
| [2] | Huang H, Sabari BR, Garcia BA, et al. SnapShot:histone modifications[J]. Cell, 2014, 159(2):458-458. e1. |
| [3] | Lawrence M, Daujat S, Schneider R. Lateral thinking:how histone modifications regulate gene expression[J]. Trends Genet, 2016, 32(1):42-56. |
| [4] | Jenuwein T, Allis CD. Translating the histone code[J]. Science, 2001, 293(5532):1074-1080. |
| [5] | Strahl BD, Allis CD. The language of covalent histone modifications[J]. Nature, 2000, 403(6765):41-45. |
| [6] | Feng Q, Wang HB, Ng HH, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain[J]. Curr Biol, 2002, 12(12):1052-1058. |
| [7] | Yeates TO. Structures of SET domain proteins:protein lysine methyltransferases make their mark[J]. Cell, 2002, 111(1):5-7. |
| [8] | Ng DWK, Wang T, Chandrasekharan MB, et al. Plant SET domain-containing proteins:structure, function and regulation[J]. Biochim Biophys Acta, 2007, 1769(5/6):316-329. |
| [9] | Ahmad A, Dong YZ, Cao XF. Characterization of the PRMT gene family in rice reveals conservation of arginine methylation[J]. PLoS One, 2011, 6(8):e22664. |
| [10] | Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation[J]. Prog Nucleic Acid Res Mol Biol, 1998, 61:65-131. |
| [11] | Boisvert FM, Chénard CA, Richard S. Protein interfaces in signaling regulated by arginine methylation[J]. Sci STKE, 2005, 2005(271):re2. |
| [12] | Bedford MT, Clarke SG. Protein arginine methylation in mammals:who, what, and why[J]. Mol Cell, 2009, 33(1):1-13. |
| [13] | Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants[J]. Curr Opin Plant Biol, 2009, 12(2):133-139. |
| [14] | Luo M, Liu XC, Singh P, et al. Chromatin modifications and remodeling in plant abiotic stress responses[J]. Biochim Biophys Acta, 2012, 1819(2):129-136. |
| [15] | Kumar SV, Wigge PA. H2A. Z-containing nucleosomes mediate the thermosensory response in Arabidopsis[J]. Cell, 2010, 140(1):136-147. |
| [16] | Ding Y, Lapko H, Ndamukong I, et al. The Arabidopsis chromatin modifier ATX1, the myotubularin-like AtMTM and the response to drought[J]. Plant Signal Behav, 2009, 4(11):1049-1058. |
| [17] | Chen K, Du KX, Shi YC, et al. H3K36 methyltransferase SDG708 enhances drought tolerance by promoting abscisic acid biosynthesis in rice[J]. New Phytol, 2021, 230(5):1967-1984. |
| [18] | Sun XW, Chen L, et al. Histone methyltransferase SDG8 in dehydr-ation stress[J]. J Univ Sci Technol China, 2021, 51(2):140-146. |
| [19] | 赵燕昊, 曹跃芬, 孙威怡, 等. 小麦抗旱研究进展[J]. 植物生理学报, 2016, 52(12):1795-1803. |
| Zhao YH, Cao YF, Sun WY, et al. The research advances in drought resistance in wheat[J]. Plant Physiol J, 2016, 52(12):1795-1803. | |
| [20] | 杨梅. 普通小麦转TaEBP基因系抗旱特性研究[D]. 杨凌: 西北农林科技大学, 2012. |
| Yang M. Study on drought-resistance of transgenic wheat with TaEBP gene[D]. Yangling: Northwest A & F University, 2012. | |
| [21] | Edae EA, Byrne PF, et al. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes[J]. Theor Appl Genet, 2014, 127(4):791-807. |
| [22] | Chen CJ, Chen H, Zhang Y, et al. TBtools:an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8):1194-1202. |
| [23] | Gasteiger E, Hoogland C, Gattiker A, et al. Protein identification and analysis tools on the ExPASy server[M]// Walker JM. The Proteomics Protocols Handbook. Totowa, NJ: Humana Press, 2005:571-607. |
| [24] | Chou KC, Shen HB. Cell-PLoc:a package of Web servers for predicting subcellular localization of proteins in various organisms[J]. Nat Protoc, 2008, 3(2):153-162. |
| [25] | Geourjon C, Deléage G. SOPMA:significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments[J]. Comput Appl Biosci, 1995, 11(6):681-684. |
| [26] | Kelley LA, Mezulis S, Yates CM, et al. The Phyre2 web portal for protein modeling, prediction and analysis[J]. Nat Protoc, 2015, 10(6):845-858. |
| [27] | Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers[J]. Proc Int Conf Intell Syst Mol Biol, 1994, 2:28-36. |
| [28] | Kumar S, Stecher G, Li M, et al. MEGA X:molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6):1547-1549. |
| [29] | He ZL, Zhang HK, Gao SH, et al. Evolview v2:an online visualization and management tool for customized and annotated phylogenetic trees[J]. Nucleic Acids Res, 2016, 44(W1):W236-W241. |
| [30] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1):325-327. |
| [31] | Ding Y, Avramova Z, Fromm M. The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways[J]. Plant J, 2011, 66(5):735-744. |
| [32] | Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat[J]. Front Plant Sci, 2014, 5:170. |
| [33] | Cao XY, Costa LM, Biderre-Petit C, et al. Abscisic acid and stress signals induce viviparous1 expression in seed and vegetative tissues of maize[J]. Plant Physiol, 2007, 143(2):720-731. |
| [34] | Yang WT, Baek D, Yun DJ, et al. Rice OsMYB5P improves plant phosphate acquisition by regulation of phosphate transporter[J]. PLoS One, 2018, 13(3):e0194628. |
| [35] | Wu JD, Jiang YL, Liang YN, et al. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants[J]. Plant Physiol Biochem, 2019, 137:179-188. |
| [36] | Xu ZJ, Sun ML, Jiang XF, et al. Glycinebetaine biosynthesis in response to osmotic stress depends on jasmonate signaling in watermelon suspension cells[J]. Front Plant Sci, 2018, 9:1469. |
| [37] | 马超, 张均, 等. 外源MeJA对花后干旱胁迫下小麦光合特性的影响[J]. 麦类作物学报, 2018, 38(5):563-571. |
| Ma C, Zhang J, et al. Effect of exogenous methyl jasmonate on photosynthetic characteristics in wheat under drought stress after anthesis[J]. J Triticeae Crops, 2018, 38(5):563-571. | |
| [38] | Rouster J, Leah R, et al. Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain[J]. Plant J, 1997, 11(3):513-523. |
| [39] | Aquea F, Vega A, Timmermann T, et al. Genome-wide analysis of the set domain group family in grapevine[J]. Plant Cell Rep, 2011, 30(6):1087-1097. |
| [40] | Napsucialy-Mendivil S, Alvarez-Venegas R, Shishkova S, et al. Arabidopsis homolog of trithorax1(ATX1)is required for cell production, patterning, and morphogenesis in root development[J]. J Exp Bot, 2014, 65(22):6373-6384. |
| [41] | Choi SC, Lee S, Kim SR, et al. Trithorax group protein Oryza sativa trithorax1 controls flowering time in rice via interaction with early heading date3[J]. Plant Physiol, 2014, 164(3):1326-1337. |
| [42] | Hang RL, Liu CY, Ahmad A, et al. Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing[J]. PNAS, 2014, 111(45):16190-16195. |
| [43] | Han YF, et al. SUVR2 is involved in transcriptional gene silencing by associating with SNF2-related chromatin-remodeling proteins in Arabidopsis[J]. Cell Res, 2014, 24(12):1445-1465. |
| [44] | Luo YX, Han YF, Zhao QY, et al. Sumoylation of SUVR2 contributes to its role in transcriptional gene silencing[J]. Sci China Life Sci, 2018, 61(2):235-243. |
| [1] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [2] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [3] | WEN Xiao-lei, LI Jian-yuan, LI Na, ZHANG Na, YANG Wen-xiang. Construction and Utilization of Yeast Two-hybrid cDNA Library of Wheat Interacted by Puccinia triticina [J]. Biotechnology Bulletin, 2023, 39(9): 136-146. |
| [4] | LIU Wen-jin, MA Rui, LIU Sheng-yan, YANG Jiang-wei, ZHANG Ning, SI Huai-jun. Cloning of StCIPK11 Gene and Analysis of Its Response to Drought Stress in Solanum tuberosum [J]. Biotechnology Bulletin, 2023, 39(9): 147-155. |
| [5] | HAN Zhi-yang, JIA Zi-miao, LIANG Qiu-ju, WANG Ke, TANG Hua-li, YE Xing-guo, ZHANG Shuang-xi. Salt Tolerance at Seedling Stage and Analysis of Selenium and Folic Acid Content in Seeds in Two Sets of Wheat-Dasypyrum villosum Chromosom Additional Lines [J]. Biotechnology Bulletin, 2023, 39(8): 185-193. |
| [6] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [7] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [8] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [9] | DING Kai-xin, WANG Li-chun, TIAN Guo-kui, WANG Hai-yan, LI Feng-yun, PAN Yang, PANG Ze, SHAN Ying. Research Progress in Uniconazole Alleviating Plant Drought Damage [J]. Biotechnology Bulletin, 2023, 39(6): 1-11. |
| [10] | LI Xin-yi, JIANG Chun-xiu, XUE Li, JIANG Hong-tao, YAO Wei, DENG Zu-hu, ZHANG Mu-qing, YU Fan. Enhancing Hybridization Signal of Sugarcane Chromosome Oligonucleotide Probe via Multiple Fluorescence Labeled Primers [J]. Biotechnology Bulletin, 2023, 39(5): 103-111. |
| [11] | WANG Chun-yu, LI Zheng-jun, WANG Ping, ZHANG Li-xia. Physiological and Biochemical Analysis of Drought Resistance in Sorghum Cuticular Wax-deficient Mutant sb1 [J]. Biotechnology Bulletin, 2023, 39(5): 160-167. |
| [12] | WANG Yi-fan, HOU Lin-hui, CHANG Yong-chun, YANG Ya-jie, CHEN Tian, ZHAO Zhu-yue, RONG Er-hua, WU Yu-xiang. Synthesis and Character Identification of Allohexaploid Between Gossypium hirsutum and G. gossypioides [J]. Biotechnology Bulletin, 2023, 39(5): 168-176. |
| [13] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [14] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [15] | HOU Xiao-yuan, CHE Zheng-zheng, LI Heng-jing, DU Chong-yu, XU Qian, WANG Qun-qing. Construction of the Soybean Membrane System cDNA Library and Interaction Proteins Screening for Effector PsAvr3a [J]. Biotechnology Bulletin, 2023, 39(4): 268-276. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||