








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (8): 118-126.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1192
Previous Articles Next Articles
HUANG Jing1( ), ZHU Liang1,2, XUE Peng-bo1,2, FU Qiang1(
), ZHU Liang1,2, XUE Peng-bo1,2, FU Qiang1( )
)
Received:2021-09-15
Online:2022-08-26
Published:2022-09-14
Contact:
FU Qiang
E-mail:jhuang@gxu.edu.cn;gxfuq@gxu.edu.cn
HUANG Jing, ZHU Liang, XUE Peng-bo, FU Qiang. Research on Mechanism and QTL Mapping Associated with Cadmium Accumulation in Rice Leaves and Grains[J]. Biotechnology Bulletin, 2022, 38(8): 118-126.
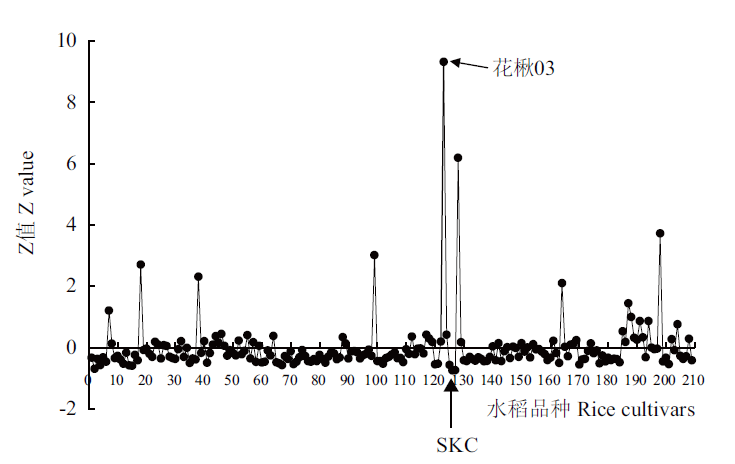
Fig. 1 Profiling of Cd accumulation in grains of rice core germplasm Z value =(individual value-population average)/population SD,and represents SD from the average value of the whole population. X axis refers to the serial number of rice cultivars
| 土壤类型Soil type | 镉Cd | 铁Fe | 锌Zn | 铜Cu | 锰Mn | 砷As |
|---|---|---|---|---|---|---|
| 对照水槽Control sink | 0.14±0.02 | 19 047.64±232.07 | 157.34±8.26 | 23.51±9.55 | 318.16±3.63 | 9.69±2.69 |
| 污染水槽 Contaminated sink | 1.09±0.11 | 20 495.83±270.84 | 271.98±9.51 | 45.39±8.31 | 322.02±9.58 | 12.99±1.28 |
| 污染大田土壤Contaminated paddy field | 0.55±0.07 | 19 603.91±267.35 | 265.89±4.15 | 48.20±3.93 | 330.8±14.11 | 12.53±1.34 |
Table 1 Contents of Cd and other metals in the soil of contaminated paddy field,contaminated sink and control sink (μg·g-1 DW)
| 土壤类型Soil type | 镉Cd | 铁Fe | 锌Zn | 铜Cu | 锰Mn | 砷As |
|---|---|---|---|---|---|---|
| 对照水槽Control sink | 0.14±0.02 | 19 047.64±232.07 | 157.34±8.26 | 23.51±9.55 | 318.16±3.63 | 9.69±2.69 |
| 污染水槽 Contaminated sink | 1.09±0.11 | 20 495.83±270.84 | 271.98±9.51 | 45.39±8.31 | 322.02±9.58 | 12.99±1.28 |
| 污染大田土壤Contaminated paddy field | 0.55±0.07 | 19 603.91±267.35 | 265.89±4.15 | 48.20±3.93 | 330.8±14.11 | 12.53±1.34 |
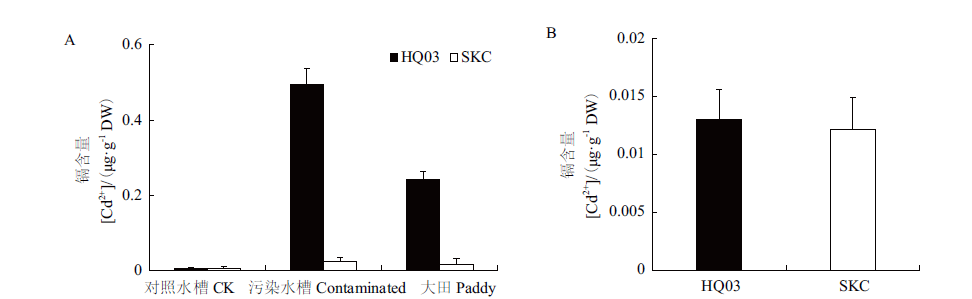
Fig.3 Cd accumulation in the grains and milled rice of HQ03 and SKC A:Cd accumulation in the grains of HQ03 and SKC from the rice grown in the contaminated paddy field,contaminated and control sink,respectively. B:Cd accumulation in the milled rice of HQ03 and SKC from the rice grown in the contaminated sink. **:P < 0.01. The same below
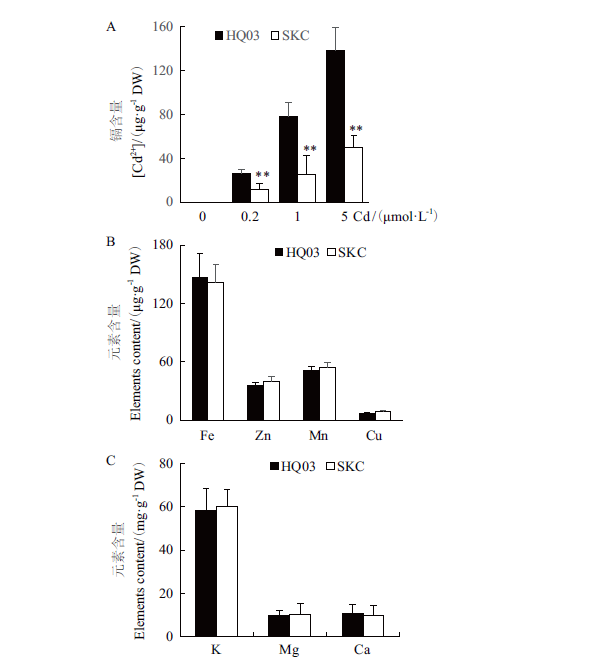
Fig.4 Elements accumulation in the leaves of HQ03 and SKC A:Cd accumulation in the leaves of HQ03 and SKC. B, C:Contents of metal elements such Fe(B)and major elements such as K(C)in the leaves of HQ03 and SKC
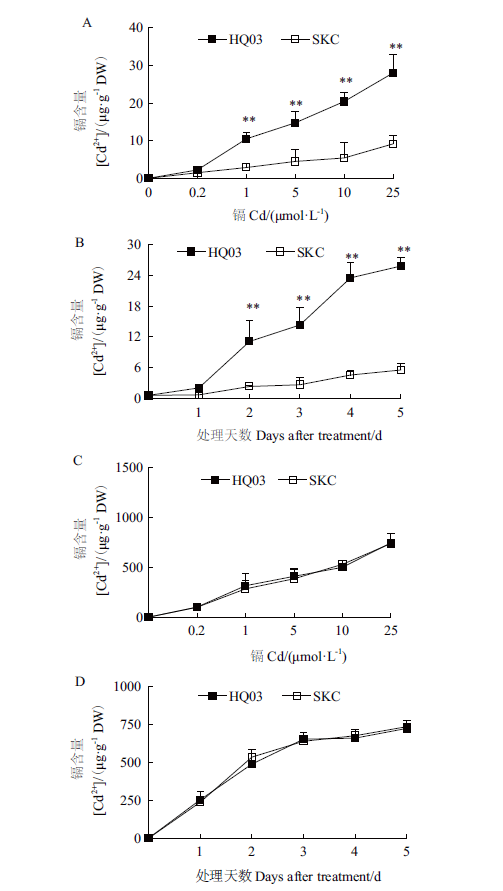
Fig.5 Cd accumulation in the leaves and roots of HQ03 and SKC from rice seedlings exposed to CdCl2 for indicated concentrations and days A:Cd levels in leaves from rice seedlings exposed to CdCl2 for 7 d at indicated concentrations;B:Cd levels in leaves from rice seedlings exposed to 10 μmol/L CdCl2 for indicated days;C:Cd levels in roots from rice seedlings exposed to CdCl2 for 7 d at indicated concentrations;D:Cd levels in roots from rice seedlings exposed to 10 μmol/L CdCl2 for indicated days
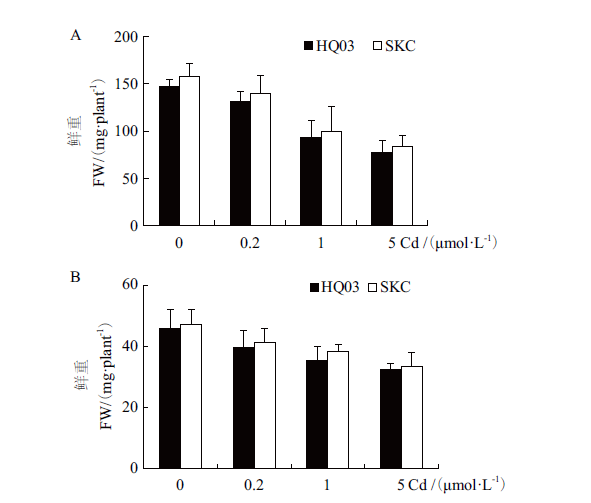
Fig. 6 Tolerance assay of HQ03 and SKC to Cd A:Fresh weight(FW)of shoots of HQ03 and SKC under different concentrations of Cd treatment. B:Fresh weight(FW)of roots of HQ03 and SKC under different concentrations of Cd treatment
| 数量性状基因座位QTL | 染色体Chr. | 标记区间Marker interval | LOD值LOD value | 贡献率 R2/% | 加性效应Additive effect |
|---|---|---|---|---|---|
| qLCd2 | 2 | RM450-RM5472 | 2.70 | 13.4 | 12.792 |
| qLCd4 | 4 | RM307-RM401 | 2.14 | 11.1 | 11.234 |
| qLCd10 | 10 | RM271-RM258 | 3.02 | 10.6 | -0.081 8 |
| qLCd11 | 11 | RM167-RM220 | 2.07 | 11.0 | 7.576 1 |
Table 2 Quantitative trait loci(QTLs)mapping for Cd accumulation in rice leaves
| 数量性状基因座位QTL | 染色体Chr. | 标记区间Marker interval | LOD值LOD value | 贡献率 R2/% | 加性效应Additive effect |
|---|---|---|---|---|---|
| qLCd2 | 2 | RM450-RM5472 | 2.70 | 13.4 | 12.792 |
| qLCd4 | 4 | RM307-RM401 | 2.14 | 11.1 | 11.234 |
| qLCd10 | 10 | RM271-RM258 | 3.02 | 10.6 | -0.081 8 |
| qLCd11 | 11 | RM167-RM220 | 2.07 | 11.0 | 7.576 1 |
| 数量性状基因座位QTL | 染色体Chr. | 标记区间Marker interval | LOD值LOD value | 贡献率R2/% | 加性效应Additive effect |
|---|---|---|---|---|---|
| qGCd2 | 2 | RM324-RM341 | 3.39 | 20.2 | -0.187 4 |
| qGCd3 | 3 | RM6266-RM2334 | 3.81 | 39.4 | 0.125 4 |
| qGCd7 | 7 | RM6574-RM6449 | 2.51 | 13.2 | -5.381 7 |
| qGCd8 | 8 | RM1235-RM1376 | 3.15 | 20.4 | -0.278 1 |
Table 3 Quantitative trait loci(QTLs)mapping for Cd accumulation in rice grains
| 数量性状基因座位QTL | 染色体Chr. | 标记区间Marker interval | LOD值LOD value | 贡献率R2/% | 加性效应Additive effect |
|---|---|---|---|---|---|
| qGCd2 | 2 | RM324-RM341 | 3.39 | 20.2 | -0.187 4 |
| qGCd3 | 3 | RM6266-RM2334 | 3.81 | 39.4 | 0.125 4 |
| qGCd7 | 7 | RM6574-RM6449 | 2.51 | 13.2 | -5.381 7 |
| qGCd8 | 8 | RM1235-RM1376 | 3.15 | 20.4 | -0.278 1 |
| [1] |
Haider FU, Liqun C, Coulter JA, et al. Cadmium toxicity in plants:Impacts and remediation strategies[J]. Ecotoxicol Environ Saf, 2021, 211:111887.
doi: 10.1016/j.ecoenv.2020.111887 URL |
| [2] | Chen HM, Chen WC, Chen YX, et al. Pollution of heavy metals in soil-plant system[M]. Beijing: Science Press, 1996. |
| [3] |
Song Y, Wang Y, Mao WF, et al. Dietary cadmium exposure assessment among the Chinese population[J]. PLoS One, 2017, 12(5):e0177978.
doi: 10.1371/journal.pone.0177978 URL |
| [4] |
Nawrot TS, Staessen JA, Roels HA, et al. Cadmium exposure in the population:from health risks to strategies of prevention[J]. Biometals, 2010, 23(5):769-782.
doi: 10.1007/s10534-010-9343-z pmid: 20517707 |
| [5] |
Meng L, Huang TH, Shi JC, et al. Decreasing cadmium uptake of rice(Oryza sativa L.)in the cadmium-contaminated paddy field through different cultivars coupling with appropriate soil amendments[J]. J Soils Sediments, 2019, 19(4):1788-1798.
doi: 10.1007/s11368-018-2186-x URL |
| [6] |
Mendoza-Cózatl DG, Jobe TO, Hauser F, et al. Long-distance transport, vacuolar sequestration, tolerance, and transcriptional responses induced by cadmium and arsenic[J]. Curr Opin Plant Biol, 2011, 14(5):554-562.
doi: 10.1016/j.pbi.2011.07.004 pmid: 21820943 |
| [7] | Zhao JL, Yang W, Zhang SH, et al. Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection[J]. Rice:N Y, 2018, 11(1):61. |
| [8] |
Xue DW, Chen MC, Zhang GP. Mapping of QTLs associated with cadmium tolerance and accumulation during seedling stage in rice(Oryza sativa L.)[J]. Euphytica, 2008, 165(3):587-596.
doi: 10.1007/s10681-008-9785-3 URL |
| [9] |
Ueno D, Kono I, Yokosho K, et al. A major quantitative trait locus controlling cadmium translocation in rice(Oryza sativa)[J]. New Phytol, 2009, 182(3):644-653.
doi: 10.1111/j.1469-8137.2009.02784.x pmid: 19309445 |
| [10] |
Ishikawa S, Abe T, Kuramata M, et al. A major quantitative trait locus for increasing cadmium-specific concentration in rice grain is located on the short arm of chromosome 7[J]. J Exp Bot, 2010, 61(3):923-934.
doi: 10.1093/jxb/erp360 pmid: 20022924 |
| [11] |
Ueno D, Yamaji N, Kono I, et al. Gene limiting cadmium accumulation in rice[J]. Proc Natl Acad Sci USA, 2010, 107(38):16500-16505.
doi: 10.1073/pnas.1005396107 URL |
| [12] |
Miyadate H, Adachi S, Hiraizumi A, et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles[J]. New Phytol, 2011, 189(1):190-199.
doi: 10.1111/j.1469-8137.2010.03459.x URL |
| [13] |
Huang Y, Sun CX, Min J, et al. Association mapping of quantitative trait loci for mineral element contents in whole grain rice(Oryza sativa L.)[J]. J Agric Food Chem, 2015, 63(50):10885-10892.
doi: 10.1021/acs.jafc.5b04932 URL |
| [14] |
Wang FJ, Wang M, Liu ZB, et al. Different responses of low grain-Cd-accumulating and high grain-Cd-accumulating rice cultivars to Cd stress[J]. Plant Physiol Biochem, 2015, 96:261-269.
doi: 10.1016/j.plaphy.2015.08.001 URL |
| [15] |
Luo JS, Huang J, Zeng DL, et al. A defensin-like protein drives cadmium efflux and allocation in rice[J]. Nat Commun, 2018, 9(1):645.
doi: 10.1038/s41467-018-03088-0 URL |
| [16] |
Hu DW, Sheng ZH, Li QL, et al. Identification of QTLs associated with cadmium concentration in rice grains[J]. J Integr Agric, 2018, 17(7):1563-1573.
doi: 10.1016/S2095-3119(17)61847-1 URL |
| [17] |
Liu WQ, Pan XW, Li YC, et al. Identification of QTLs and validation of qCd-2 associated with grain cadmium concentrations in rice[J]. Rice Sci, 2019, 26(1):42-49.
doi: 10.1016/j.rsci.2018.12.003 URL |
| [18] |
Yan HL, Xu WX, Xie JY, et al. Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies[J]. Nat Commun, 2019, 10(1):2562.
doi: 10.1038/s41467-019-10544-y URL |
| [19] |
Pan XW, Li YC, Liu WQ, et al. QTL mapping and candidate gene analysis of cadmium accumulation in polished rice by genome-wide association study[J]. Sci Rep, 2020, 10(1):11791.
doi: 10.1038/s41598-020-68742-4 URL |
| [20] | Lincoln SE, Daly MJ, Lander ES. Constructing genetic linkage maps with mapmaker/Exp version 3.0: A tutorial and reference manual[M]. 3rd ed. Cambridge: Whitehead Institute for Biometrical Research, 1993. |
| [21] | Lincoln SE, Daly MJ, Lander ES. Mapping genes controlling quantitative traits using MAPMAKER/QTL version 1.1: A tutorial and reference manual[M]. 2nd ed. Cambridge: Whitehead Institute for Biometrical Research, 1993. |
| [22] |
Uraguchi S, Mori S, Kuramata M, et al. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice[J]. J Exp Bot, 2009, 60(9):2677-2688.
doi: 10.1093/jxb/erp119 pmid: 19401409 |
| [23] |
Takahashi A, Agrawal GK, Yamazaki M, et al. Rice Pti1a negatively regulates RAR1-dependent defense responses[J]. Plant Cell, 2007, 19(9):2940-2951.
pmid: 17890377 |
| [1] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [2] | WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq [J]. Biotechnology Bulletin, 2023, 39(8): 173-184. |
| [3] | YAO Sha-sha, WANG Jing-jing, WANG Jun-jie, LIANG Wei-hong. Molecular Mechanisms of Rice Grain Size Regulation Related to Plant Hormone Signaling Pathways [J]. Biotechnology Bulletin, 2023, 39(8): 80-90. |
| [4] | XU Jian-xia, DING Yan-qing, FENG Zhou, CAO Ning, CHENG Bin, GAO Xu, ZOU Gui-hua, ZHANG Li-yi. QTL Mapping of Sorghum Plant Height and Internode Numbers Based on Super-GBS Technique [J]. Biotechnology Bulletin, 2023, 39(7): 185-194. |
| [5] | YU Hui, WANG Jing, LIANG Xin-xin, XIN Ya-ping, ZHOU Jun, ZHAO Hui-jun. Isolation and Functional Verification of Genes Responding to Iron and Cadmium Stresses in Lycium barbarum [J]. Biotechnology Bulletin, 2023, 39(7): 195-205. |
| [6] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [7] | LIANG Cheng-gang, WANG Yan, LI Tian, OHSUGI Ryu, AOKI Naohiro. Effect of SP1 on Panicle Architecture by Regulating Carbohydrate Remobilization [J]. Biotechnology Bulletin, 2023, 39(5): 152-159. |
| [8] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [9] | ZHOU Ding-ding, LI Hui-hu, TANG Xing-yong, YU Fa-xin, KONG Dan-yu, LIU Yi. Research Progress in the Biosynthesis and Regulation of Glycyrrhizic Acid and Liquiritin [J]. Biotechnology Bulletin, 2023, 39(5): 44-53. |
| [10] | YANG Mao, LIN Yu-feng, DAI Yang-shuo, PAN Su-jun, PENG Wei-ye, YAN Ming-xiong, LI Wei, WANG Bing, DAI Liang-ying. OsDIS1 Negatively Regulates Rice Drought Tolerance Through Antioxidant Pathways [J]. Biotechnology Bulletin, 2023, 39(2): 88-95. |
| [11] | JIANG Min-xuan, LI Kang, LUO Liang, LIU Jian-xiang, LU Hai-ping. Advances on the Expressions of Foreign Proteins in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 110-122. |
| [12] | JIANG Nan, SHI Yang, ZHAO Zhi-hui, LI Bin, ZHAO Yi-hui, YANG Jun-biao, YAN Jia-ming, JIN Yu-fan, CHEN Ji, HUANG Jin. Expression and Functional Analysis of OsPT1 Gene in Rice Under Cadmium Stress [J]. Biotechnology Bulletin, 2023, 39(1): 166-174. |
| [13] | CHEN Guang, LI Jia, DU Rui-ying, WANG Xu. Identification and Gene Functional Analysis of Salinity-hypersensitive Mutant ss2 in Rice [J]. Biotechnology Bulletin, 2022, 38(9): 158-166. |
| [14] | GAO Xiao-rong, DING Yao, LV Jun. Effects of Pseudomonas sp. PR3,a Pyrene-degrading Bacterium with Plant Growth-promoting Properties,on Rice Growth Under Pyrene Stress [J]. Biotechnology Bulletin, 2022, 38(9): 226-236. |
| [15] | CHEN Guang, LI Jia, DU Rui-ying, WANG Xu. pOsHAK1:OsFLN2 Expression Enhances the Drought Tolerance by Altering Sugar Metabolism in Rice [J]. Biotechnology Bulletin, 2022, 38(8): 92-100. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||