








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (8): 198-205.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1482
Previous Articles Next Articles
TANG Guang-fu( ), GUI Yan-ling, MAN Hai-qiao, ZHAO Jie-hong(
), GUI Yan-ling, MAN Hai-qiao, ZHAO Jie-hong( )
)
Received:2021-11-29
Online:2022-08-26
Published:2022-09-14
Contact:
ZHAO Jie-hong
E-mail:1838921871@qq.com;105798711@qq.com
TANG Guang-fu, GUI Yan-ling, MAN Hai-qiao, ZHAO Jie-hong. Editing pyrG Gene of Monascus by CRISPR/Cas 9 and Its Effects on Secondary Metabolism[J]. Biotechnology Bulletin, 2022, 38(8): 198-205.
| 引物名称 Primer name | 引物或sgRNA序列 Primer or sgRNA sequence(5'-3') | 退火温度 Annea-ling temperature/℃ | 靶标序列长度 Target sequence length/bp |
|---|---|---|---|
| Cas9-F | GTTTAAGGTCCTGGGCAACA | 55 | 312 |
| Cas9-R | ATCGTGGGGTACTTCTCGTG | ||
| PyrGgr-F | GTTGAAACGAACCCCAGCAC | 57 | 380 |
| pyrGgr-R | CCACATCGACATCCTCTCCG | ||
| sgRNA | TTAATACGACTCACTATAGGGggcttgaagttcctgcgttgGTTTTAGAGCTAGAAATA | / | / |
Table 1 Primers and sgRNA sequences for PCR detection
| 引物名称 Primer name | 引物或sgRNA序列 Primer or sgRNA sequence(5'-3') | 退火温度 Annea-ling temperature/℃ | 靶标序列长度 Target sequence length/bp |
|---|---|---|---|
| Cas9-F | GTTTAAGGTCCTGGGCAACA | 55 | 312 |
| Cas9-R | ATCGTGGGGTACTTCTCGTG | ||
| PyrGgr-F | GTTGAAACGAACCCCAGCAC | 57 | 380 |
| pyrGgr-R | CCACATCGACATCCTCTCCG | ||
| sgRNA | TTAATACGACTCACTATAGGGggcttgaagttcctgcgttgGTTTTAGAGCTAGAAATA | / | / |

Fig. 3 Transformation of Cas 9 into monascus mediated by A. tumefaciens A:PCR detection of A. tumefaciens. B:Co-culture of A. tumefaciens and monascus. C:PCR detection of the Cas 9 of monascus chassis strain. 1-8:Different strains. M:DL1000 DNA marker
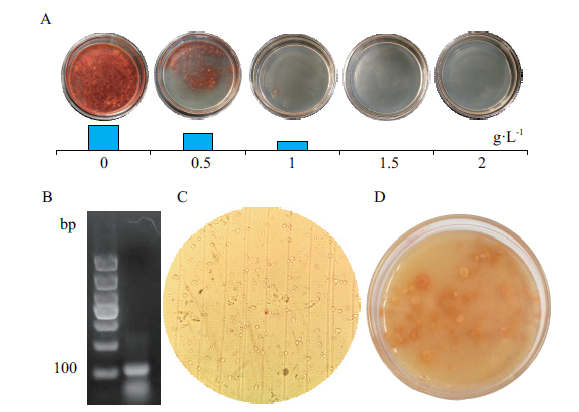
Fig. 4 Transformation of monascus protoplasts by sgRNA in vitro A:Inhibition of 5-FOA on monascus. B:Electrophoresis of the sgRNA. C:Prepared protoplasts. D:Screening resistant monascus
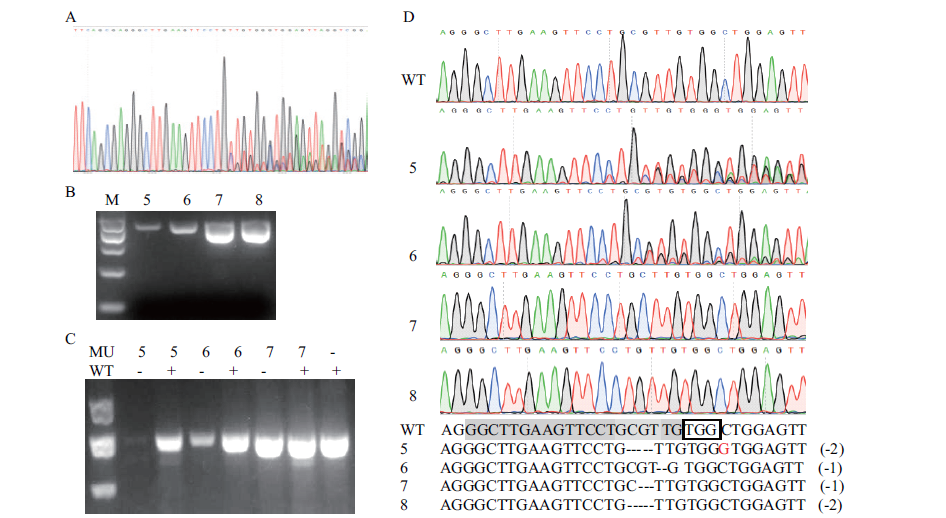
Fig. 5 Variation analysis of pyrG gene of monascus mutant A:Part nested peak sequence. B:PCR detection of pyrG gene in some strains. C:PCR product cutted by T7EI. D:Mutation loci of pyrG gene in some strains. MU:Mutant. WT:Wild-type. “+”:With DNA. “-”:Without DNA. Black box:Refers to the PAM site. Grey background:Refers to the sgRNA target sequence
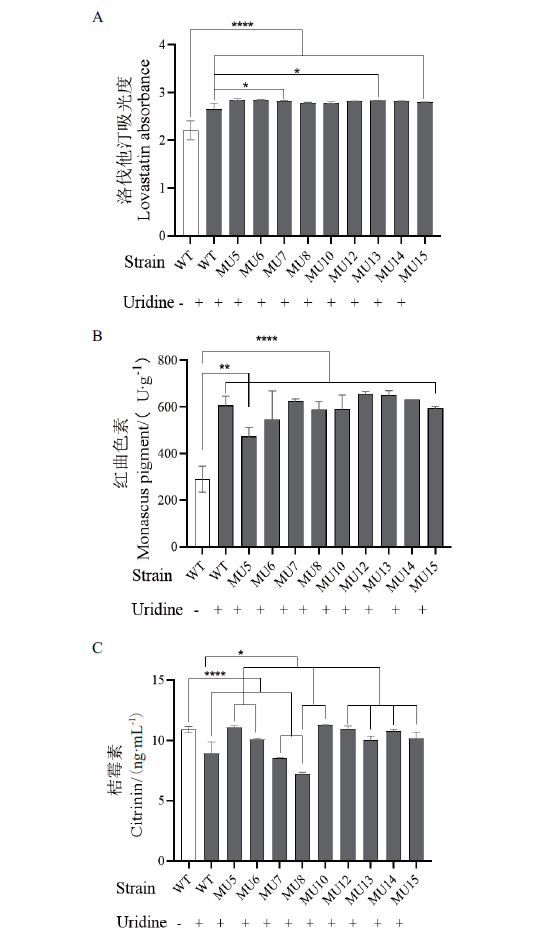
Fig. 7 Effect of uracil riboside on the chemical composition content of different strains in medium A:Lovastatin absorbance. B:Monascus pigment content. C:Citrinin content. “-”:Not added. “+”:Added. WT:Wild type strain. MU:Mutant strain
| [1] | 傅金泉. 中国红曲及其实用技术[M]. 北京: 中国轻工业出版社, 1997. |
| Fu JQ. Chinese red yeast rice and its practical technology[M]. Beijing: China Light Industry Press, 1997. | |
| [2] | 李利, 陈莎, 陈福生, 等. 红曲菌次生代谢产物生物合成途径及相关基因的研究进展[J]. 微生物学通报, 2013, 40(2):294-303. |
| Li L, Chen S, Chen FS, et al. Review on biosynthetic pathway of secondary metabolites and the related genes in Monascus spp[J]. Microbiol China, 2013, 40(2):294-303. | |
| [3] |
Blanc PJ, Laussac JP, Bars JL, et al. Characterization of monascidin A from Monascus as citrinin[J]. Int J Food Microbiol, 1995, 27(2/3):201-213.
doi: 10.1016/0168-1605(94)00167-5 URL |
| [4] | 李贞景, 薛意斌, 刘妍, 等. 红曲菌中桔霉素的控制策略及研究进展[J]. 食品科学, 2018, 39(17):263-268. |
| Li ZJ, Xue YB, Liu Y, et al. Recent progress on control strategies against citrinin in Monascus spp[J]. Food Sci, 2018, 39(17):263-268. | |
| [5] |
Shimizu T, Kinoshita H, Nihira T. Identification and in vivo functional analysis by gene disruption of ctnA, an activator gene involved in citrinin biosynthesis in Monascus purpureus[J]. Appl Environ Microbiol, 2007, 73(16):5097-5103.
doi: 10.1128/AEM.01979-06 URL |
| [6] |
Li YP, Pan YF, Zou LH, et al. Lower citrinin production by gene disruption of ctnB involved in citrinin biosynthesis in Monascus aurantiacus Li AS3. 4384[J]. J Agric Food Chem, 2013, 61(30):7397-7402.
doi: 10.1021/jf400879s URL |
| [7] |
Ning ZQ, Cui H, Xu Y, et al. Deleting the citrinin biosynthesis-related gene, ctnE, to greatly reduce citrinin production in Monascus aurantiacus Li AS3. 4384[J]. Int J Food Microbiol, 2017, 241:325-330.
doi: 10.1016/j.ijfoodmicro.2016.11.004 URL |
| [8] | Li YP, Wang N, Jiao XX, et al. The ctnF gene is involved in citrinin and pigment synthesis in Monascus aurantiacus[J]. J Basic Microbiol, 2020, 60(10):873-881. |
| [9] |
Li YP, Tang X, Wu W, et al. The ctnG gene encodes carbonic anhydrase involved in mycotoxin citrinin biosynthesis from Monascus aurantiacus[J]. Food Addit Contam Part A Chem Anal Control Expo Risk Assess, 2015, 32(4):577-583.
doi: 10.1080/19440049.2014.990993 URL |
| [10] | 吴伟. 橙色红曲菌ctnG基因和ctnH基因缺失菌株的构建及其功能分析[D]. 南昌: 南昌大学, 2010. |
| Wu W. Construction and functional analysis of the ctnG and ctnH gene disruption mutants in Monascus aurantiacus AS3. 4384[D]. Nanchang: Nanchang University, 2010. | |
| [11] |
Balakrishnan B, Chandran R, Park SH, et al. Delineating citrinin biosynthesis:Ctn-ORF3 dioxygenase-mediated multi-step methyl oxidation precedes a reduction-mediated pyran ring cyclization[J]. Bioorg Med Chem Lett, 2016, 26(2):392-396.
doi: S0960-894X(15)30314-0 pmid: 26707397 |
| [12] | 崔华. 橙色红曲菌桔霉素合成相关基因-orf3和ctnE缺失菌株的构建及其相关分析[D]. 南昌: 南昌大学, 2012. |
| Cui H. Construction and correlation analysis of the Orf3and ctnE gene disruption mutants in Monascus aurantiacus AS3. 4384[D]. Nanchang: Nanchang University, 2012. | |
| [13] |
Liang B, Du XJ, Li P, et al. Orf6 gene encoded glyoxalase involved in mycotoxin citrinin biosynthesis in Monascus purpureus YY-1[J]. Appl Microbiol Biotechnol, 2017, 101(19):7281-7292.
doi: 10.1007/s00253-017-8462-7 pmid: 28831532 |
| [14] | 邹乐花, 李燕萍, 黄志兵, 等. 橙色红曲菌As3. 4384 orf7基因缺失株的构建及其功能分析[J]. 中国生物工程杂志, 2011, 31(7):79-84. |
| Zou LH, Li YP, Huang ZB, et al. Construction of the Monascus aurantiacus As3. 4384 orf7 gene deletion strains and functional analysis[J]. China Biotechnol, 2011, 31(7):79-84. | |
| [15] |
Shimizu T, Kinoshita H, Ishihara S, et al. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus[J]. Appl Environ Microbiol, 2005, 71(7):3453-3457.
doi: 10.1128/AEM.71.7.3453-3457.2005 URL |
| [16] |
Arbour CA, Imperiali B. Uridine natural products:challenging targets and inspiration for novel small molecule inhibitors[J]. Bioorg Med Chem, 2020, 28(18):115661.
doi: 10.1016/j.bmc.2020.115661 URL |
| [17] |
Zhang LH, Zheng XM, Cairns TC, et al. Disruption or reduced expression of the orotidine-5'-decarboxylase gene pyrG increases citric acid production:a new discovery during recyclable genome editing in Aspergillus niger[J]. Microb Cell Fact, 2020, 19(1):76.
doi: 10.1186/s12934-020-01334-z URL |
| [18] | 王汝毅. 根癌农杆菌介导红曲霉转化库的构建及转化子性质研究[D]. 武汉: 华中农业大学, 2005. |
| Wang RY. Construction of transformation library of Monascus ruber mediated by Agrobacterium tumefaciens and study on transformants’ characters[D]. Wuhan: Huazhong Agricultural University, 2005. | |
| [19] | 周礼红, 李国琴, 王正祥, 等. 红曲霉原生质体的制备、再生及其遗传转化系统[J]. 遗传, 2005, 27(3):423-428. |
| Zhou LH, Li GQ, Wang ZX, et al. Preparation and regeneration of protoplasts from Monascus purpureus and genetic transformation system[J]. Hered Beijing, 2005, 27(3):423-428. | |
| [20] | 李亮. 高产洛伐他汀红曲菌株的选育及发酵条件研究[D]. 福州: 福州大学, 2016. |
| Li L. Research of breeding and fermentation conditions of high-lovastatin Monascus strains[D]. Fuzhou: Fuzhou University, 2016. | |
| [21] |
Liu R, Chen L, Jiang YP, et al. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system[J]. Cell Discov, 2015, 1:15007.
doi: 10.1038/celldisc.2015.7 URL |
| [22] |
Shen B, Zhang J, Wu HY, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting[J]. Cell Res, 2013, 23(5):720-723.
doi: 10.1038/cr.2013.46 pmid: 23545779 |
| [23] |
Song RJ, Zhai Q, Sun L, et al. CRISPR/Cas9 genome editing technology in filamentous fungi:progress and perspective[J]. Appl Microbiol Biotechnol, 2019, 103(17):6919-6932.
doi: 10.1007/s00253-019-10007-w URL |
| [24] |
O’Donovan GA, Neuhard J. Pyrimidine metabolism in microorganisms[J]. Bacteriol Rev, 1970, 34(3):278-343.
doi: 10.1128/br.34.3.278-343.1970 URL |
| [1] | PAN Guo-qiang, WU Si-yuan, LIU Lu, GUO Hui-ming, CHENG Hong-mei, SU Xiao-feng. Construction and Preliminary Analysis of Verticillim dahliae Mutant Library [J]. Biotechnology Bulletin, 2023, 39(5): 112-119. |
| [2] | SHI Jia, ZHU Xiu-mei, XUE Meng-yu, YU Chao, WEI Yi-ming, YANG Feng-huan, CHEN Hua-min. Optimization and Application of the Chromatin Immunoprecipitation Based on Rice Protoplast [J]. Biotechnology Bulletin, 2022, 38(7): 62-69. |
| [3] | LIANG Ling, HUANG Qin-geng, WENG Xue-qing, WU Song-gang, HUANG Jian-zhong. Breeding L-Glutamic Acid Producing Engineering Strain by Mutagenesis and Its Fermentation Efficiency [J]. Biotechnology Bulletin, 2020, 36(6): 143-149. |
| [4] | PAN Xu-yao, WEI Tao, TAN Zhu-hao, LIN Long-zhen, GUO Li-qiong, LIN Jun-fang. Breeding of a New Strain of High-yield Surfactin by Bacillus Interspecies Protoplast Fusion [J]. Biotechnology Bulletin, 2019, 35(8): 238-245. |
| [5] | ZHAO Xiao-qiang, CHEN Zhi-rong, HE Fang, SHEN Nan, GAO Feng, HUANG Jia-feng. Preparation and Regeneration of Protoplast from Verticillium dahliae [J]. Biotechnology Bulletin, 2018, 34(7): 166-173. |
| [6] | SUN Xiao-rui, CHEN Bo-wen, ZHANG Xiao-lin, MENG Jun-long. Optimization of Preparation Conditions of Pleurotus geesterani Monocaryon Mycelia Protoplast [J]. Biotechnology Bulletin, 2018, 34(4): 70-76. |
| [7] | WANG Wei-wei, XIAO Yan, ZHANG Yi-xi, LIU Jing, TANG Wei, LI Can-hui. Preparation and Regeneration Method of Phytophthora infestans Protoplast in Potato [J]. Biotechnology Bulletin, 2018, 34(4): 77-82. |
| [8] | ZHANG Xiao-hui HAN Rong. Subcellular Localization of Profilin-1 from Arabidopsis Utilizing Two Transient Expression Systems [J]. Biotechnology Bulletin, 2017, 33(5): 57-62. |
| [9] | CHEN Nan, YU Fei, HE Yan-liu, BU Ning. Labeling and Tracing of Green Fluorescent Protein in Fungal Endophyte with Growth-promoting Activity to Rice Seedlings [J]. Biotechnology Bulletin, 2017, 33(3): 100-105. |
| [10] | LIN Yan-mei, LI Rui-jie, ZHANG Hui-jie, QIN Xiu-lin, FENG Jia-xun. Formation and Transformation of Protoplast of Trichoderma atroviride HP35-3 Highly Yielding Cellulase [J]. Biotechnology Bulletin, 2016, 32(9): 225-231. |
| [11] | ZHOU Ming-ming, LI Xiao-yan, REN Meng-nan, CHENG Ke-meng, HUANG Hai-dong. Conditions for Protoplast Preparation and Regeneration by Sphingomonas sp. ATCC 31555 [J]. Biotechnology Bulletin, 2016, 32(7): 126-130. |
| [12] | XI Hai-xiu, AI Ke-jun, TONG Shao-ming. The Optimization of Isolation Method for Mesophyll Protoplast from Common Wheat(Triticum aestivum)Seedlings [J]. Biotechnology Bulletin, 2016, 32(4): 68-73. |
| [13] | GOU Min, YANG Bai-xue, TANG Yue-qin, Kida Kenji. Construction of Acid-tolerant and Flocculating Saccharomyces cerevisiae Strain for Ethanol Production by Protoplast Fusion [J]. Biotechnology Bulletin, 2016, 32(11): 115-123. |
| [14] | Hu Jiya, Lu Ping, Niu Yanfang. Preparation and Regeneration of Protoplasts Isolated from Embellisia Fungal Endophyte of Oxytropis glabra [J]. Biotechnology Bulletin, 2015, 31(5): 134-139. |
| [15] | Zhang Xiaoli, Zheng Xiaomei, Man Yun, Luo Hu, Yu Jiandong, Zheng Ping, Liu Hao, Sun Jibin. Preparation of Protoplast for Efficient DNA Transformation of Citric Acid Hyper-producing Aspergillus niger Industrial Strain [J]. Biotechnology Bulletin, 2015, 31(3): 171-177. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||