








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (9): 116-126.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1544
Previous Articles Next Articles
HU Xue-ying1,2( ), ZHANG Yue2, GUO Ya-jie2, QIU Tian-lei2, GAO Min2, SUN Xing-bin1(
), ZHANG Yue2, GUO Ya-jie2, QIU Tian-lei2, GAO Min2, SUN Xing-bin1( ), WANG Xu-ming2(
), WANG Xu-ming2( )
)
Received:2021-12-14
Online:2022-09-26
Published:2022-10-11
Contact:
SUN Xing-bin,WANG Xu-ming
E-mail:17745161087@163.com;sunxingbin1025@163.com;wangxuming@baafs.net.cn
HU Xue-ying, ZHANG Yue, GUO Ya-jie, QIU Tian-lei, GAO Min, SUN Xing-bin, WANG Xu-ming. Comparison in Antibiotic Resistance Genes Carried by Bacteriophages and Bacteria in Farmland Soil Amended with Different Fertilizers[J]. Biotechnology Bulletin, 2022, 38(9): 116-126.
| 处理Treatment | 施肥量Fertilizer amount/(kg· m-2) |
|---|---|
| 不施肥(CK) Without fertilizer | 0 |
| 单施化肥(IF) Inorganic fertilizer amendment | Urea 0.04、(NH4)2HPO4 0.05和K2SO4 0.03 |
| 单施有机肥(OF) Organic fertilizer amendment | Compost from chicken manure 2.38 |
Table 1 Fertilizer amount of different treatments
| 处理Treatment | 施肥量Fertilizer amount/(kg· m-2) |
|---|---|
| 不施肥(CK) Without fertilizer | 0 |
| 单施化肥(IF) Inorganic fertilizer amendment | Urea 0.04、(NH4)2HPO4 0.05和K2SO4 0.03 |
| 单施有机肥(OF) Organic fertilizer amendment | Compost from chicken manure 2.38 |
| 抗生素抗性基因ARGs | 引物序列Primer sequences | 退火温度Annealing temperature Tm/oC | 参考文献Reference | |
|---|---|---|---|---|
| 氨基糖苷类 Aminoglycosides | strA | CCGGTGGCATTTGAGAAAAA | 60 | [ |
| GTGGCTCAACCTGCGAAAAG | ||||
| strB | GCTCGGTCGTGAGAACAATCT | 60 | [ | |
| CAATTTCGGTCGCCTGGTAGT | ||||
| aadA-01 | GTTGTGCACGACGACATCATT | 60 | [ | |
| GGCTCGAAGATACCTGCAAGAA | ||||
| β-内酰胺类 β-lactams | blaOXA-20 | TGATGATTGTCGAAGCCAAA | 60 | [ |
| GCCTGTAGGCCACTCTACCC | ||||
| blaTEM | AGCATCTTACGGATGGCATGA | 60 | [ | |
| TCCTCCGATCGTTGTCAGAAGT | ||||
| blaCTX-M | CAGATTCGGTTCGCTTTCAC | 55 | [ | |
| GCAAATACTTTATCGTGCTGATG | ||||
| 大环内酯-林肯酰胺-链阳性菌素B MLSB | ermA | TTGAGAAGGGATTTGCGAAAAG | 60 | [ |
| ATATCCATCTCCACCATTAATAGTAAACC | ||||
| ermB | TAAAGGGCATTTAACGACGAAACT | 60 | [ | |
| TTTATACCTCTGTTTGTTAGGGAATTGAA | ||||
| mphA-01 | CTGACGCGCTCCGTGTT | 60 | [ | |
| GGTGGTGCATGGCGATCT | ||||
| oleC | CCCGGAGTCGATGTTCGA | 60 | [ | |
| GCCGAAGACGTACACGAACAG | ||||
| 磺胺类 Sulfonamides | sul1 | CAGCGCTATGCGCTCAAG | 60 | [ |
| ATCCCGCTGCGCTGAGT | ||||
| sul2 | TCATCTGCCAAACTCGTCGTTA | 60 | [ | |
| GTCAAAGAACGCCGCAATGT | ||||
| 四环素类Tetracyclines | tetA | CTCACCAGCCTGACCTCGAT | 60 | [ |
| CACGTTGTTATAGAAGCCGCATAG | ||||
| tetW | ATGAACATTCCCACCGTTATCTTT | 60 | [ | |
| ATATCGGCGGAGAGCTTATCC | ||||
| tetM | GGAGCGATTACAGAATTAGGAAGC | 60 | [ | |
| TCCATATGTCCTGGCGTGTC | ||||
| tetX | AAATTTGTTACCGACACGGAAGTT | 60 | [ | |
| CATAGCTGAAAAAATCCAGGACAGTT | ||||
| 多耐药类 Multi-drug resistance | acrA-05 | CGTGCGCGAACGAACA | 60 | [ |
| ACTTTGCGCGCCATCTTC | ||||
| emrD | CTCAGCAGTATGGTGGTAAGCATT | 60 | [ | |
| ACCAGGCGCCGAAGAAC | ||||
| mepA | ATCGGTCGCTCTTCGTTCAC | 60 | [ | |
| ATAAATAGGATCGAGCTGCTGGAT | ||||
| mexF | CCGCGAGAAGGCCAAGA | 60 | [ | |
| TTGAGTTCGGCGGTGATGA | ||||
| 喹诺酮类 Quinolones | qnrA | AGGATTTCTCACGCCAGGATT | 60 | [ |
| CCGCTTTCAATGAAACTGCAA | ||||
| qnrS | CGACGTGCTAACTTGCGTGA | 60 | [ | |
| GGCATTGTTGGAAACTTGCA | ||||
| 万古霉素类 Vancomycins | vanHB | GAGGTTTCCGAGGCGACAA | 60 | [ |
| CTCTCGGCGGCAGTCGTAT | ||||
| vanA | GGGCTGTGAGGTCGGTTG | 60 | [ | |
| TTCAGTACAATGCGGCCGTTA | ||||
| 多黏菌素类 Polymyxins | mcr-1 | CACATCGACGGCGTATTCTG | 60 | [ |
| CAACGAGCATACCGACATCG | ||||
| 可移动遗传元件 MGEs | intl1 | CCGTAGAACAAGCAGGCATCA | 55 | [ |
| GCGTTGAAATCATCGTCGTAGAG | ||||
Table 2 PCR primer information of ARGs and class I integrase gene(intl1)
| 抗生素抗性基因ARGs | 引物序列Primer sequences | 退火温度Annealing temperature Tm/oC | 参考文献Reference | |
|---|---|---|---|---|
| 氨基糖苷类 Aminoglycosides | strA | CCGGTGGCATTTGAGAAAAA | 60 | [ |
| GTGGCTCAACCTGCGAAAAG | ||||
| strB | GCTCGGTCGTGAGAACAATCT | 60 | [ | |
| CAATTTCGGTCGCCTGGTAGT | ||||
| aadA-01 | GTTGTGCACGACGACATCATT | 60 | [ | |
| GGCTCGAAGATACCTGCAAGAA | ||||
| β-内酰胺类 β-lactams | blaOXA-20 | TGATGATTGTCGAAGCCAAA | 60 | [ |
| GCCTGTAGGCCACTCTACCC | ||||
| blaTEM | AGCATCTTACGGATGGCATGA | 60 | [ | |
| TCCTCCGATCGTTGTCAGAAGT | ||||
| blaCTX-M | CAGATTCGGTTCGCTTTCAC | 55 | [ | |
| GCAAATACTTTATCGTGCTGATG | ||||
| 大环内酯-林肯酰胺-链阳性菌素B MLSB | ermA | TTGAGAAGGGATTTGCGAAAAG | 60 | [ |
| ATATCCATCTCCACCATTAATAGTAAACC | ||||
| ermB | TAAAGGGCATTTAACGACGAAACT | 60 | [ | |
| TTTATACCTCTGTTTGTTAGGGAATTGAA | ||||
| mphA-01 | CTGACGCGCTCCGTGTT | 60 | [ | |
| GGTGGTGCATGGCGATCT | ||||
| oleC | CCCGGAGTCGATGTTCGA | 60 | [ | |
| GCCGAAGACGTACACGAACAG | ||||
| 磺胺类 Sulfonamides | sul1 | CAGCGCTATGCGCTCAAG | 60 | [ |
| ATCCCGCTGCGCTGAGT | ||||
| sul2 | TCATCTGCCAAACTCGTCGTTA | 60 | [ | |
| GTCAAAGAACGCCGCAATGT | ||||
| 四环素类Tetracyclines | tetA | CTCACCAGCCTGACCTCGAT | 60 | [ |
| CACGTTGTTATAGAAGCCGCATAG | ||||
| tetW | ATGAACATTCCCACCGTTATCTTT | 60 | [ | |
| ATATCGGCGGAGAGCTTATCC | ||||
| tetM | GGAGCGATTACAGAATTAGGAAGC | 60 | [ | |
| TCCATATGTCCTGGCGTGTC | ||||
| tetX | AAATTTGTTACCGACACGGAAGTT | 60 | [ | |
| CATAGCTGAAAAAATCCAGGACAGTT | ||||
| 多耐药类 Multi-drug resistance | acrA-05 | CGTGCGCGAACGAACA | 60 | [ |
| ACTTTGCGCGCCATCTTC | ||||
| emrD | CTCAGCAGTATGGTGGTAAGCATT | 60 | [ | |
| ACCAGGCGCCGAAGAAC | ||||
| mepA | ATCGGTCGCTCTTCGTTCAC | 60 | [ | |
| ATAAATAGGATCGAGCTGCTGGAT | ||||
| mexF | CCGCGAGAAGGCCAAGA | 60 | [ | |
| TTGAGTTCGGCGGTGATGA | ||||
| 喹诺酮类 Quinolones | qnrA | AGGATTTCTCACGCCAGGATT | 60 | [ |
| CCGCTTTCAATGAAACTGCAA | ||||
| qnrS | CGACGTGCTAACTTGCGTGA | 60 | [ | |
| GGCATTGTTGGAAACTTGCA | ||||
| 万古霉素类 Vancomycins | vanHB | GAGGTTTCCGAGGCGACAA | 60 | [ |
| CTCTCGGCGGCAGTCGTAT | ||||
| vanA | GGGCTGTGAGGTCGGTTG | 60 | [ | |
| TTCAGTACAATGCGGCCGTTA | ||||
| 多黏菌素类 Polymyxins | mcr-1 | CACATCGACGGCGTATTCTG | 60 | [ |
| CAACGAGCATACCGACATCG | ||||
| 可移动遗传元件 MGEs | intl1 | CCGTAGAACAAGCAGGCATCA | 55 | [ |
| GCGTTGAAATCATCGTCGTAGAG | ||||
| 抗生素抗性基因亚型 ARG subtypes | 细菌Bacteria | 噬菌体acteriophages | ||||||
|---|---|---|---|---|---|---|---|---|
| CK | IF | OF | CK | IF | OF | |||
| strA | + | + | + | - | + | + | ||
| strB | + | + | + | + | + | + | ||
| aadA-01 | + | + | + | + | + | + | ||
| blaOXA-20 | - | - | - | - | - | - | ||
| blaTEM | + | + | + | + | + | + | ||
| blaCTX-M | - | + | + | - | - | + | ||
| ermA | + | + | + | - | - | - | ||
| ermB | + | + | + | - | - | - | ||
| mphA-01 | + | + | + | + | + | + | ||
| oleC | + | + | + | + | + | + | ||
| sul1 | + | + | + | - | - | - | ||
| sul2 | + | + | + | + | + | + | ||
| tetA | + | + | + | + | + | + | ||
| tetW | + | + | + | + | + | + | ||
| tetM | + | + | + | + | + | + | ||
| tetX | + | + | + | + | + | + | ||
| acrA-05 | + | + | + | + | + | + | ||
| emrD | + | + | + | + | + | + | ||
| mepA | + | + | + | + | + | + | ||
| mexF | + | + | + | + | + | + | ||
| qnrA | - | - | - | - | - | - | ||
| qnrS | + | + | + | - | + | - | ||
| vanHB | + | + | + | + | + | + | ||
| vanA | + | + | + | + | + | + | ||
| mcr-1 | + | + | + | + | - | + | ||
| 检出率Detection rate/% | 88 | 92 | 92 | 68 | 72 | 76 | ||
| intl1 | + | + | + | + | + | + | ||
Table 3 ARGs and intl1 carried by bacteriophages and bacteria in farmland soil under different ferti-lization treatments
| 抗生素抗性基因亚型 ARG subtypes | 细菌Bacteria | 噬菌体acteriophages | ||||||
|---|---|---|---|---|---|---|---|---|
| CK | IF | OF | CK | IF | OF | |||
| strA | + | + | + | - | + | + | ||
| strB | + | + | + | + | + | + | ||
| aadA-01 | + | + | + | + | + | + | ||
| blaOXA-20 | - | - | - | - | - | - | ||
| blaTEM | + | + | + | + | + | + | ||
| blaCTX-M | - | + | + | - | - | + | ||
| ermA | + | + | + | - | - | - | ||
| ermB | + | + | + | - | - | - | ||
| mphA-01 | + | + | + | + | + | + | ||
| oleC | + | + | + | + | + | + | ||
| sul1 | + | + | + | - | - | - | ||
| sul2 | + | + | + | + | + | + | ||
| tetA | + | + | + | + | + | + | ||
| tetW | + | + | + | + | + | + | ||
| tetM | + | + | + | + | + | + | ||
| tetX | + | + | + | + | + | + | ||
| acrA-05 | + | + | + | + | + | + | ||
| emrD | + | + | + | + | + | + | ||
| mepA | + | + | + | + | + | + | ||
| mexF | + | + | + | + | + | + | ||
| qnrA | - | - | - | - | - | - | ||
| qnrS | + | + | + | - | + | - | ||
| vanHB | + | + | + | + | + | + | ||
| vanA | + | + | + | + | + | + | ||
| mcr-1 | + | + | + | + | - | + | ||
| 检出率Detection rate/% | 88 | 92 | 92 | 68 | 72 | 76 | ||
| intl1 | + | + | + | + | + | + | ||
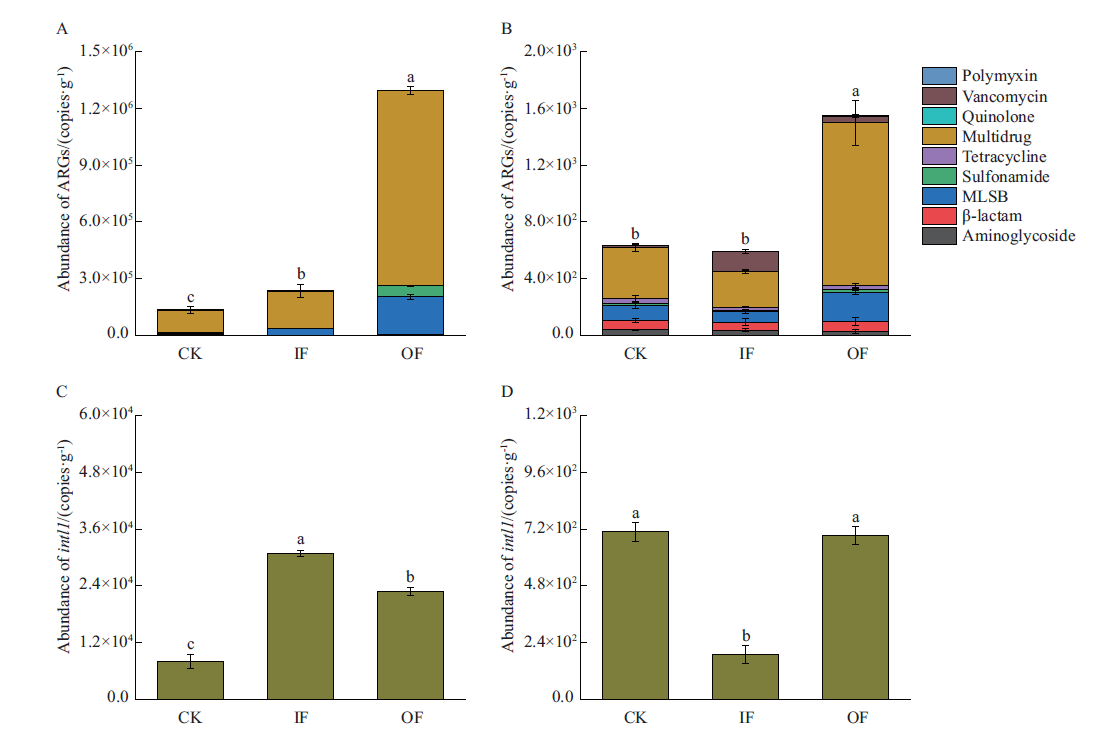
Fig. 1 Abundances of ARGs and intl1 in bacteria(A,C)and bacteriophages(B,D)in farmland soil under different fertilization treatments Different lowercase letters indicate the statistically significant difference(P<0.05). The same below
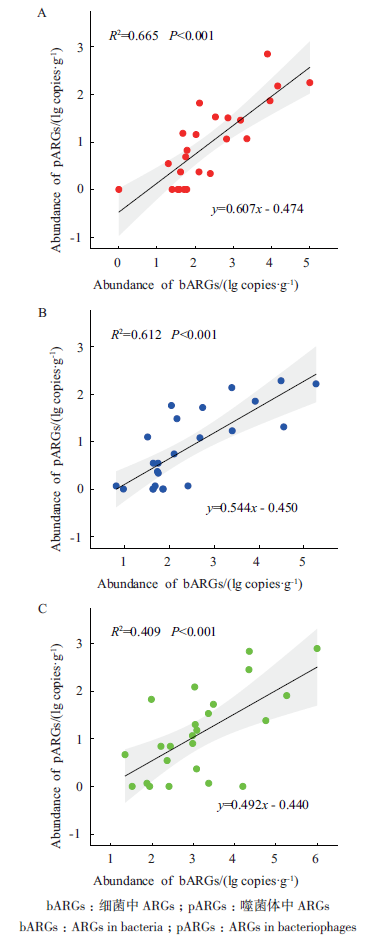
Fig. 3 Correlation of the abundances of ARGs between bacteria and bacteriophages fractions in farmland soil under different fertilization treatments(A:CK;B:IF;C:OF)
| [1] |
Pruden A, Pei RT, Storteboom H, et al. Antibiotic resistance genes as emerging contaminants:studies in northern Colorado[J]. Environ Sci Technol, 2006, 40(23):7445-7450.
pmid: 17181002 |
| [2] | 朱冬, 陈青林, 丁晶, 等. 土壤生态系统中抗生素抗性基因与星球健康:进展与展望[J]. 中国科学:生命科学, 2019, 49(12):1652-1663. |
|
Zhu D, Chen QL, Ding J, et al. Antibiotic resistance genes in the soil ecosystem and planetary health:Progress and prospect[J]. Sci Sin Vitae, 2019, 49(12):1652-1663.
doi: 10.1360/SSV-2019-0267 URL |
|
| [3] |
D'Costa VM, King CE, Kalan L, et al. Antibiotic resistance is ancient[J]. Nature, 2011, 477(7365):457-461.
doi: 10.1038/nature10388 URL |
| [4] |
Zhu YG, Johnson TA, Su JQ, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms[J]. PNAS, 2013, 110(9):3435-3440.
doi: 10.1073/pnas.1222743110 URL |
| [5] |
Udikovic-Kolic N, Wichmann F, Broderick NA, et al. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization[J]. Proc Natl Acad Sci USA, 2014, 111(42):15202-15207.
doi: 10.1073/pnas.1409836111 URL |
| [6] |
Xie WY, Shen Q, Zhao FJ. Antibiotics and antibiotic resistance from animal manures to soil:a review[J]. Eur J Soil Sci, 2018, 69(1):181-195.
doi: 10.1111/ejss.12494 URL |
| [7] |
Balcázar JL. How do bacteriophages promote antibiotic resistance in the environment?[J]. Clin Microbiol Infect, 2018, 24(5):447-449.
doi: 10.1016/j.cmi.2017.10.010 URL |
| [8] |
Muniesa M, Colomer-Lluch M, Jofre J. Potential impact of environmental bacteriophages in spreading antibiotic resistance genes[J]. Future Microbiol, 2013, 8(6):739-751.
doi: 10.2217/fmb.13.32 pmid: 23701331 |
| [9] |
Dion MB, Oechslin F, Moineau S. Phage diversity, genomics and phylogeny[J]. Nat Rev Microbiol, 2020, 18(3):125-138.
doi: 10.1038/s41579-019-0311-5 pmid: 32015529 |
| [10] |
Yang YX, Shi WJ, Lu SY, et al. Prevalence of antibiotic resistance genes in bacteriophage DNA fraction from Funan River water in Sichuan, China[J]. Sci Total Environ, 2018, 626:835-841.
doi: 10.1016/j.scitotenv.2018.01.148 URL |
| [11] |
Larrañaga O, Brown-Jaque M, Quirós P, et al. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil[J]. Environ Int, 2018, 115:133-141.
doi: S0160-4120(18)30048-5 pmid: 29567433 |
| [12] |
Sun MM, Ye M, Jiao WT, et al. Changes in tetracycline partitioning and bacteria/phage-comediated ARGs in microplastic-contaminated greenhouse soil facilitated by sophorolipid[J]. J Hazard Mater, 2018, 345:131-139.
doi: S0304-3894(17)30858-0 pmid: 29175125 |
| [13] |
Wang MZ, Liu P, Zhou Q, et al. Estimating the contribution of bacteriophage to the dissemination of antibiotic resistance genes in pig feces[J]. Environ Pollut, 2018, 238:291-298.
doi: S0269-7491(17)34918-7 pmid: 29573711 |
| [14] |
Calero-Cáceres W, Ye M, Balcázar JL. Bacteriophages as environmental reservoirs of antibiotic resistance[J]. Trends Microbiol, 2019, 27(7):570-577.
doi: S0966-842X(19)30059-9 pmid: 30905524 |
| [15] |
Oliveira J, Mahony J, Hanemaaijer L, et al. Detecting Lactococcus lactis prophages by mitomycin C-mediated induction coupled to flow cytometry analysis[J]. Front Microbiol, 2017, 8:1343.
doi: 10.3389/fmicb.2017.01343 pmid: 28769907 |
| [16] | Lekunberri I, Subirats J, Borrego CM, et al. Exploring the contribution of bacteriophages to antibiotic resistance[J]. Environ Pollut, 2017, 220(Pt B):981-984. |
| [17] |
Ouyang WY, Huang FY, Zhao Y, et al. Increased levels of antibiotic resistance in urban stream of Jiulongjiang River, China[J]. Appl Microbiol Biotechnol, 2015, 99(13):5697-5707.
doi: 10.1007/s00253-015-6416-5 URL |
| [18] |
Bert F, Branger C, Lambert-Zechovsky N. Identification of PSE and OXA beta-lactamase genes in Pseudomonas aeruginosa using PCR-restriction fragment length polymorphism[J]. J Antimicrob Chemother, 2002, 50(1):11-18.
doi: 10.1093/jac/dkf069 URL |
| [19] |
Chen ZY, Zhang W, Yang LX, et al. Antibiotic resistance genes and bacterial communities in cornfield and pasture soils receiving swine and dairy manures[J]. Environ Pollut, 2019, 248:947-957.
doi: S0269-7491(18)34775-4 pmid: 30861417 |
| [20] |
Colomer-Lluch M, Jofre J, Muniesa M. Quinolone resistance genes(qnrA and qnrS)in bacteriophage particles from wastewater samples and the effect of inducing agents on packaged antibiotic resistance genes[J]. J Antimicrob Chemother, 2014, 69(5):1265-1274.
doi: 10.1093/jac/dkt528 pmid: 24458509 |
| [21] |
Kenzaka T, Tani K, Sakotani A, et al. High-frequency phage-mediated gene transfer among Escherichia coli cells, determined at the single-cell level[J]. Appl Environ Microbiol, 2007, 73(10):3291-3299.
doi: 10.1128/AEM.02890-06 URL |
| [22] |
Subirats J, Sànchez-Melsió A, Borrego CM, et al. Metagenomic analysis reveals that bacteriophages are reservoirs of antibiotic resistance genes[J]. Int J Antimicrob Agents, 2016, 48(2):163-167.
doi: 10.1016/j.ijantimicag.2016.04.028 URL |
| [23] | 黄福义, 周曙仡聃, 王佳妮, 等. 不同作物农田土壤抗生素抗性基因多样性[J]. 环境科学, 2021, 42(6):2975-2980. |
| Huang FY, Zhou S, Wang JN, et al. Profiling of antibiotic resistance genes in different croplands[J]. Environ Sci, 2021, 42(6):2975-2980. | |
| [24] |
Forsberg KJ, Patel S, Gibson MK, et al. Bacterial phylogeny structures soil resistomes across habitats[J]. Nature, 2014, 509(7502):612-616.
doi: 10.1038/nature13377 URL |
| [25] |
Sui QW, Zhang JY, Chen MX, et al. Fate of microbial pollutants and evolution of antibiotic resistance in three types of soil amended with swine slurry[J]. Environ Pollut, 2019, 245:353-362.
doi: S0269-7491(18)32705-2 pmid: 30448505 |
| [26] |
Debroas D, Siguret C. Viruses as key reservoirs of antibiotic resistance genes in the environment[J]. ISME J, 2019, 13(11):2856-2867.
doi: 10.1038/s41396-019-0478-9 pmid: 31358910 |
| [27] |
Lood R, Ertürk G, Mattiasson B. Revisiting antibiotic resistance spreading in wastewater treatment plants - bacteriophages as a much neglected potential transmission vehicle[J]. Front Microbiol, 2017, 8:2298.
doi: 10.3389/fmicb.2017.02298 pmid: 29209304 |
| [28] |
Anand T, Bera BC, Vaid RK, et al. Abundance of antibiotic resistance genes in environmental bacteriophages[J]. J Gen Virol, 2016, 97(12):3458-3466.
doi: 10.1099/jgv.0.000639 pmid: 27902329 |
| [29] |
Ross J, Topp E. Abundance of antibiotic resistance genes in bacteriophage following soil fertilization with dairy manure or municipal biosolids, and evidence for potential transduction[J]. Appl Environ Microbiol, 2015, 81(22):7905-7913.
doi: 10.1128/AEM.02363-15 URL |
| [30] |
Yang YX, Xie XJ, Tang MJ, et al. Exploring the profile of antimicrobial resistance genes harboring by bacteriophage in chicken feces[J]. Sci Total Environ, 2020, 700:134446.
doi: 10.1016/j.scitotenv.2019.134446 URL |
| [31] |
Brown-Jaque M, Calero-Cáceres W, Espinal P, et al. Antibiotic resistance genes in phage particles isolated from human faeces and induced from clinical bacterial isolates[J]. Int J Antimicrob Agents, 2018, 51(3):434-442.
doi: 10.1016/j.ijantimicag.2017.11.014 URL |
| [32] |
Calero-Cáceres W, Muniesa M. Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater[J]. Water Res, 2016, 95:11-18.
doi: 10.1016/j.watres.2016.03.006 pmid: 26978717 |
| [33] | Yang YW, Xing SC, Chen YX, et al. Profiles of bacteria/phage-comediated ARGs in pig farm wastewater treatment plants in China:association with mobile genetic elements, bacterial communities and environmental factors[J]. J Hazard Mater, 2021, 404(Pt B):124149. |
| [34] |
Dickinson AW, Power A, Hansen MG, et al. Heavy metal pollution and co-selection for antibiotic resistance:a microbial palaeontology approach[J]. Environ Int, 2019, 132:105117.
doi: 10.1016/j.envint.2019.105117 URL |
| [35] | Zhao ZL, Wang J, Han Y, et al. Nutrients, heavy metals and microbial communities co-driven distribution of antibiotic resistance genes in adjacent environment of mariculture[J]. Environ Pollut, 2017, 220(Pt B):909-918. |
| [1] | CHENG Shen-wei, ZHANG Ke-qiang, LIANG Jun-feng, LIU Fu-yuan, GAO Xing-liang, DU Lian-zhu. Establishment of a Triple Droplet Digital PCR Quantitative Detection Method for Typical Pathogenic Bacteria in Livestock and Poultry Manure [J]. Biotechnology Bulletin, 2022, 38(9): 271-280. |
| [2] | WANG Yong, LAN Qing-kuo, ZHAO Xin, CHEN Rui, SHEN Xiao-ling, LI Wen, ZHANG Yao-zhong, LI Liang, WANG Qin-ying. Estimation of the Copy Number of Exogenous Genes in Genetically Modified Rice by Droplet Digital PCR [J]. Biotechnology Bulletin, 2018, 34(3): 53-58. |
| [3] | LIU Xiu-xia, XU Hai-yan, XIN Guo-qin, MU Xi-jun, SUN Xue-sen, GU Wei. Biological Characteristics of Bacillus subtilis Bacteriophage and Mutagenesis Breeding of Resistant Strains [J]. Biotechnology Bulletin, 2017, 33(2): 143-148. |
| [4] | LI Ming-yuan1, 2, WANG Ji-lian1, REN Yu1, JI Xiu-ling3, Gulbahar·Sawut1. Biological Characteristics of Three Pseudomonas Cold-active Bacteriophages [J]. Biotechnology Bulletin, 2016, 32(9): 123-130. |
| [5] | REN Yi-fei, GAO Qin, DENG Ting-ting, LI Xiang, HUANG Wen-sheng, CHEN Shun-sheng, CHEN Ying. Establishment of Precisely Quantitative Method of Genetically Modified Rice LL62 Based on Digital PCR [J]. Biotechnology Bulletin, 2016, 32(8): 69-76. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||