








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (1): 48-58.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0053
Previous Articles Next Articles
LI Jian-jian1( ), HE Chen-jing2, HUANG Xiao-ping2(
), HE Chen-jing2, HUANG Xiao-ping2( ), XIANG Tai-he2(
), XIANG Tai-he2( )
)
Received:2022-01-12
Online:2023-01-26
Published:2023-02-02
Contact:
HUANG Xiao-ping,XIANG Tai-he
E-mail:l13849447174@163.com;xphuang_hznu@163.com;xthcn@163.com
LI Jian-jian, HE Chen-jing, HUANG Xiao-ping, XIANG Tai-he. Research Progress in the Regulation of Development and Stress Response by Long Non-coding RNAs in Plants[J]. Biotechnology Bulletin, 2023, 39(1): 48-58.
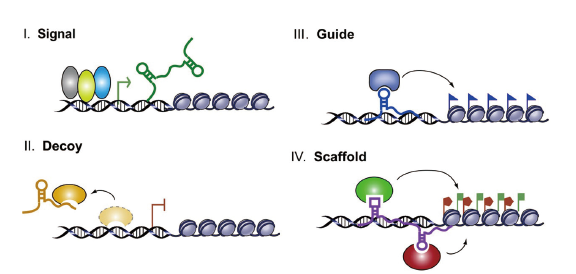
Fig. 1 Schematic diagram of the four archetypes of lncRNA mechanism Archetype I:As signals,lncRNA expression can faithfully reflect the combinatorial actions of transcription factors(colored ovals)or signaling pathways to indicate gene regulation in space and time. Archetype II:As decoys,lncRNAs can titrate transcription factors and other proteins away from chromatin. Archetype III:As guides,lncRNAs can recruit chromatin-modifying enzymes to target genes. Archetype IV:As scaffolds,lncRNAs can bring together multiple proteins to form ribonucleoprotein complexes
| 植物Plant | 长链非编码RNAs(lncRNAs) | 生物学功能Biological functions |
|---|---|---|
| 拟南芥Arabidopsis thaliana | MAS[ | 开花Flowering |
| ENOD40[ | 侧根发育Lateral root development | |
| HID1[ | 光形态发生Photomorphogenesis | |
| npc536[ | 盐胁迫Salt stress | |
| asDOG1[ | 种子萌发Seed germination | |
| IPS1[ | 磷胁迫Phosphorus stress | |
| T5120[ | 氮素胁迫Nitrogen stress | |
| HAL6[ | 温度胁迫Temperature stress | |
| lincRNA13853[ | 干旱胁迫Drought stress | |
| AtR8[ | 低氧和水杨酸胁迫low oxygen and SA stress | |
| TE-lincRNA11195[ | 脱落酸应答Abscisic acid response | |
| APOLO[ | 生长素应答Auxin response | |
| TAR-197/212[ | 病原菌应答Pathogen response | |
| 水稻Oryza sativa | ENOD40[ | 根发育Root development |
| TL[ | 叶形态发育Leaf morphological development | |
| LDMAR[ | 光敏育性Photoperiod-sensitive male sterility | |
| XLOC_057324[ | 开花Flowering | |
| EF-cd[ | 籽粒成熟Grain maturation | |
| LAIR[ | 水稻产量Rice yield | |
| MISSEN[ | 胚乳发育Endosperm development | |
| ALEX1[ | 病原菌应答Pathogen response | |
| 番茄Solanum lycopersicum | lncRNA1459[ | 果实成熟Fruit ripening |
| lncRNA16397[ | 病原菌应答Pathogen response | |
| 棉花Gossypium hirsutum | Ghir_A01G011040[ | 纤维发育Fiber development |
| XLOC_409583[ | 株高Plant height | |
| LncRNA973[ | 盐胁迫Salt stress | |
| GhlncNAT-ANX2、GhlncNAT-RLP7[ | 疾病防御Disease resistance | |
| 杨树Populus L. | lnc12、lncWOX11[ | 不定根发育Adventitious root development |
| 白菜Brassica rapa | BcMF11[ | 花粉发育Pollen development |
| 大豆Glycine max | ENOD40[ | 根发育Root development |
| 小麦Triticum aestivum | VAS[ | 开花Flowering |
| 蒺藜苜蓿Medicago truncatula | ENOD40[ | 根发育Root development |
| Mt4[ | 盐胁迫Salt stress | |
| 苹果Malus domestica | MSTRG.85814[ | 营养胁迫Nutrition stress |
| 玉米Zea mays | Zm401[ | 花粉发育Pollen development |
| GARR2[ | 激素应答Hormone response | |
| 密瓜Cucumis melo | LNC-003610[ | 果实成熟Fruit ripening |
| 苎麻Boehmeria nivea | LTCONS_00034183[ | 纤维发育Fiber development |
| 柽柳Tamarix hispida | THSAIR6[ | 盐胁迫Salt stress |
Table 1 Functional validated lncRNAs involving in plant development and stress response
| 植物Plant | 长链非编码RNAs(lncRNAs) | 生物学功能Biological functions |
|---|---|---|
| 拟南芥Arabidopsis thaliana | MAS[ | 开花Flowering |
| ENOD40[ | 侧根发育Lateral root development | |
| HID1[ | 光形态发生Photomorphogenesis | |
| npc536[ | 盐胁迫Salt stress | |
| asDOG1[ | 种子萌发Seed germination | |
| IPS1[ | 磷胁迫Phosphorus stress | |
| T5120[ | 氮素胁迫Nitrogen stress | |
| HAL6[ | 温度胁迫Temperature stress | |
| lincRNA13853[ | 干旱胁迫Drought stress | |
| AtR8[ | 低氧和水杨酸胁迫low oxygen and SA stress | |
| TE-lincRNA11195[ | 脱落酸应答Abscisic acid response | |
| APOLO[ | 生长素应答Auxin response | |
| TAR-197/212[ | 病原菌应答Pathogen response | |
| 水稻Oryza sativa | ENOD40[ | 根发育Root development |
| TL[ | 叶形态发育Leaf morphological development | |
| LDMAR[ | 光敏育性Photoperiod-sensitive male sterility | |
| XLOC_057324[ | 开花Flowering | |
| EF-cd[ | 籽粒成熟Grain maturation | |
| LAIR[ | 水稻产量Rice yield | |
| MISSEN[ | 胚乳发育Endosperm development | |
| ALEX1[ | 病原菌应答Pathogen response | |
| 番茄Solanum lycopersicum | lncRNA1459[ | 果实成熟Fruit ripening |
| lncRNA16397[ | 病原菌应答Pathogen response | |
| 棉花Gossypium hirsutum | Ghir_A01G011040[ | 纤维发育Fiber development |
| XLOC_409583[ | 株高Plant height | |
| LncRNA973[ | 盐胁迫Salt stress | |
| GhlncNAT-ANX2、GhlncNAT-RLP7[ | 疾病防御Disease resistance | |
| 杨树Populus L. | lnc12、lncWOX11[ | 不定根发育Adventitious root development |
| 白菜Brassica rapa | BcMF11[ | 花粉发育Pollen development |
| 大豆Glycine max | ENOD40[ | 根发育Root development |
| 小麦Triticum aestivum | VAS[ | 开花Flowering |
| 蒺藜苜蓿Medicago truncatula | ENOD40[ | 根发育Root development |
| Mt4[ | 盐胁迫Salt stress | |
| 苹果Malus domestica | MSTRG.85814[ | 营养胁迫Nutrition stress |
| 玉米Zea mays | Zm401[ | 花粉发育Pollen development |
| GARR2[ | 激素应答Hormone response | |
| 密瓜Cucumis melo | LNC-003610[ | 果实成熟Fruit ripening |
| 苎麻Boehmeria nivea | LTCONS_00034183[ | 纤维发育Fiber development |
| 柽柳Tamarix hispida | THSAIR6[ | 盐胁迫Salt stress |
| [1] |
Morris KV, Mattick JS. The rise of regulatory RNA[J]. Nat Rev Genet, 2014, 15(6): 423-437.
doi: 10.1038/nrg3722 pmid: 24776770 |
| [2] |
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs[J]. Cell, 2009, 136(4): 629-641.
doi: 10.1016/j.cell.2009.02.006 pmid: 19239885 |
| [3] |
Zhao XY, Li JR, Lian B, et al. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA[J]. Nat Commun, 2018, 9(1): 5056.
doi: 10.1038/s41467-018-07500-7 URL |
| [4] |
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs[J]. Mol Cell, 2011, 43(6): 904-914.
doi: 10.1016/j.molcel.2011.08.018 pmid: 21925379 |
| [5] |
Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA[J]. Science, 2011, 331(6013): 76-79.
doi: 10.1126/science.1197349 pmid: 21127216 |
| [6] |
Ding JH, Lu Q, Ouyang YD, et al. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice[J]. Proc Natl Acad Sci USA, 2012, 109(7): 2654-2659.
doi: 10.1073/pnas.1121374109 pmid: 22308482 |
| [7] |
Salmena L, Poliseno L, Tay Y, et al. A CeRNA hypothesis:the Rosetta stone of a hidden RNA language?[J]. Cell, 2011, 146(3): 353-358.
doi: 10.1016/j.cell.2011.07.014 URL |
| [8] |
Franco-Zorrilla JM, Valli A, Todesco M, et al. Target mimicry provides a new mechanism for regulation of microRNA activity[J]. Nat Genet, 2007, 39(8): 1033-1037.
doi: 10.1038/ng2079 pmid: 17643101 |
| [9] |
Campalans A, Kondorosi A, Crespi M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula[J]. Plant Cell, 2004, 16(4): 1047-1059.
pmid: 15037734 |
| [10] | 刘思安. 杨树不定根发育相关IncRNA的鉴定与功能研究[D]. 南京: 南京林业大学, 2019. |
| Liu SA. Identification and functional study of lncRNA related to adventitious root development of poplar[D]. Nanjing: Nanjing Forestry University, 2019. | |
| [11] |
Bardou F, Ariel F, Simpson CG, et al. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis[J]. Dev Cell, 2014, 30(2): 166-176.
doi: 10.1016/j.devcel.2014.06.017 URL |
| [12] |
Yang WC, Katinakis P, Hendriks P, et al. Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development[J]. Plant J, 1993, 3(4): 573-585.
doi: 10.1046/j.1365-313x.1993.03040573.x pmid: 8220464 |
| [13] |
Kouchi H, Takane K, So RB, et al. Rice ENOD40:isolation and expression analysis in rice and transgenic soybean root nodules[J]. Plant J, 1999, 18(2): 121-129.
doi: 10.1046/j.1365-313x.1999.00432.x pmid: 10363365 |
| [14] |
Campalans A, Kondorosi A, Crespi M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula[J]. Plant Cell, 2004, 16(4): 1047-1059.
pmid: 15037734 |
| [15] |
Crespi MD, Jurkevitch E, Poiret M, et al. enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth[J]. EMBO J, 1994, 13(21): 5099-5112.
doi: 10.1002/j.1460-2075.1994.tb06839.x pmid: 7957074 |
| [16] | 饶晶. 苎麻纤维发育相关的长链非编码RNA分析[D]. 北京: 中国农业科学院, 2021. |
| Rao J. Analysis of long non-coding RNA related to fiber development in ramie[D]. Beijing: Chinese Academy of Agricultural Sciences, 2021. | |
| [17] | 闫飞林. 棉花纤维起始相关长链非编码RNA的功能鉴定[D]. 武汉: 华中农业大学, 2019. |
| Yan FL. The functional identification of long non-coding RNA related to cotton fiber initial[D]. Wuhan: Huazhong Agricultural University, 2019. | |
| [18] |
Zhao T, Tao XY, Feng SL, et al. LncRNAs in polyploid cotton interspecific hybrids are derived from transposon neofunctionalization[J]. Genome Biol, 2018, 19(1): 195.
doi: 10.1186/s13059-018-1574-2 pmid: 30419941 |
| [19] | 蔺珍. 红苞凤梨lncRNAs的鉴定及lncABCG11的功能研究[D]. 雅安: 四川农业大学, 2019. |
| Lin Z. Identification of A. comosus var. bracteatus LncRNAs and functional research of lncABCG11[D]. Ya'an: Sichuan Agricultural University, 2019. | |
| [20] |
Wang YQ, Fan XD, Lin F, et al. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light[J]. Proc Natl Acad Sci USA, 2014, 111(28): 10359-10364.
doi: 10.1073/pnas.1409457111 URL |
| [21] |
Liu X, Li DY, Zhang DL, et al. A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice[J]. New Phytol, 2018, 218(2): 774-788.
doi: 10.1111/nph.15023 URL |
| [22] |
Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering[J]. Plant Cell, 1999, 11(5): 949-956.
doi: 10.1105/tpc.11.5.949 pmid: 10330478 |
| [23] |
Swiezewski S, Liu FQ, Magusin A, et al. Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target[J]. Nature, 2009, 462(7274): 799-802.
doi: 10.1038/nature08618 URL |
| [24] |
Kim DH, Sung S. Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs[J]. Dev Cell, 2017, 40(3): 302-312. e4.
doi: 10.1016/j.devcel.2016.12.021 URL |
| [25] |
Shin JH, Chekanova JA. Arabidopsis RRP6L1 and RRP6L2 function in FLOWERING LOCUS C silencing via regulation of antisense RNA synthesis[J]. PLoS Genet, 2014, 10(9): e1004612.
doi: 10.1371/journal.pgen.1004612 URL |
| [26] |
Ben Amor B, Wirth S, Merchan F, et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses[J]. Genome Res, 2009, 19(1): 57-69.
doi: 10.1101/gr.080275.108 URL |
| [27] |
Henriques R, Wang H, Liu J, et al. The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering[J]. New Phytol, 2017, 216(3): 854-867.
doi: 10.1111/nph.14703 pmid: 28758689 |
| [28] |
Xu SJ, Dong Q, Deng M, et al. The vernalization-induced long non-coding RNA VAS functions with the transcription factor TaRF2b to promote TaVRN1 expression for flowering in hexaploid wheat[J]. Mol Plant, 2021, 14(9): 1525-1538.
doi: 10.1016/j.molp.2021.05.026 URL |
| [29] |
Fan YR, Yang JY, Mathioni SM, et al. PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice[J]. Proc Natl Acad Sci USA, 2016, 113(52): 15144-15149.
doi: 10.1073/pnas.1619159114 pmid: 27965387 |
| [30] |
Zhang YC, Liao JY, Li ZY, et al. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice[J]. Genome Biol, 2014, 15(12): 512.
doi: 10.1186/s13059-014-0512-1 URL |
| [31] |
Song JH, Cao JS, Wang CG. BcMF11, a novel non-coding RNA gene from Brassica campestris, is required for pollen development and male fertility[J]. Plant Cell Rep, 2013, 32(1): 21-30.
doi: 10.1007/s00299-012-1337-6 URL |
| [32] |
Song JH, Cao JS, Yu XL, et al. BcMF11, a putative pollen-specific non-coding RNA from Brassica campestris ssp. chinensis[J]. J Plant Physiol, 2007, 164(8): 1097-1100.
doi: 10.1016/j.jplph.2006.10.002 URL |
| [33] | 崔洁. 白菜长链非编码RNA-BrLMaP的功能鉴定[D]. 杭州: 浙江大学, 2018. |
| Cui J. The functional characterization of BrLMaP, a long non-coding RNA in Brassica rapa[D]. Hangzhou: Zhejiang University, 2018. | |
| [34] |
Huang L, Dong H, Zhou D, et al. Systematic identification of long non-coding RNAs during pollen development and fertilization in Brassica rapa[J]. Plant J, 2018, 96(1): 203-222.
doi: 10.1111/tpj.14016 URL |
| [35] | Dai XY, Yu JJ, Zhao Q, et al. Non-coding RNA for ZM401, a pollen-specific gene of Zea mays[J]. Acta Bot Sin, 2004, 46(4): 497-504. |
| [36] |
Li R, Fu DQ, Zhu BZ, et al. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening[J]. Plant J, 2018, 9(3): 513-524.
doi: 10.1046/j.1365-313X.1996.09040513.x URL |
| [37] |
Zhu BZ, Yang YF, Li R, et al. RNA sequencing and functional analysis implicate the regulatory role of long non-coding RNAs in tomato fruit ripening[J]. J Exp Bot, 2015, 66(15): 4483-4495.
doi: 10.1093/jxb/erv203 pmid: 25948705 |
| [38] |
Fang J, Zhang FT, Wang HR, et al. Ef-cd locus shortens rice maturity duration without yield penalty[J]. Proc Natl Acad Sci USA, 2019, 116(37): 18717-18722.
doi: 10.1073/pnas.1815030116 pmid: 31451662 |
| [39] |
Wang Y, Luo XJ, Sun F, et al. Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice[J]. Nat Commun, 2018, 9(1): 3516.
doi: 10.1038/s41467-018-05829-7 URL |
| [40] | 田芸芸. 甜瓜果实发育相关长链非编码RNA的鉴定及调控作用研究[D]. 呼和浩特: 内蒙古大学, 2020. |
| Tian YY. Identification and regulating role study of long non-coding RNA related to the fruit development of Cucumis melo[D]. Hohhot: Inner Mongolia University, 2020. | |
| [41] |
Huang XP, Zhang HY, Wang Q, et al. Genome-wide identification and characterization of long non-coding RNAs involved in flag leaf senescence of rice[J]. Plant Mol Biol, 2021, 105(6): 655-684.
doi: 10.1007/s11103-021-01121-3 pmid: 33569692 |
| [42] | Fedak H, Palusinska M, Krzyczmonik K, et al. Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript[J]. Proc Natl Acad Sci USA, 2016, 113(48): E7846-E7855. |
| [43] |
Zhou YF, Zhang YC, Sun YM, et al. The parent-of-origin lncRNA MISSEN regulates rice endosperm development[J]. Nat Commun, 2021, 12(1): 6525.
doi: 10.1038/s41467-021-26795-7 URL |
| [44] |
Jabnoune M, Secco D, Lecampion C, et al. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness[J]. Plant Cell, 2013, 25(10): 4166-4182.
doi: 10.1105/tpc.113.116251 URL |
| [45] |
Burleigh SM, Harrison MJ. Characterization of the Mt4 gene from Medicago truncatula[J]. Gene, 1998, 216(1): 47-53.
pmid: 9714729 |
| [46] |
Burleigh SH, Harrison MJ. The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots[J]. Plant Physiol, 1999, 119(1): 241-248.
pmid: 9880366 |
| [47] |
Zhang XP, Dong J, Deng FN, et al. The long non-coding RNA lncRNA973 is involved in cotton response to salt stress[J]. BMC Plant Biol, 2019, 19(1): 459.
doi: 10.1186/s12870-019-2088-0 URL |
| [48] |
Liu F, Xu YR, Chang KX, et al. The long noncoding RNA T5120 regulates nitrate response and assimilation in Arabidopsis[J]. New Phytol, 2019, 224(1): 117-131.
doi: 10.1111/nph.16038 URL |
| [49] | 许欣, 卢惠君, 王玉成, 等. 刚毛柽柳SAIR6长链非编码RNA耐盐功能初探[J]. 北京林业大学学报, 2021, 43(3): 36-43. |
| Xu X, Lu HJ, Wang YC, et al. Salt stress tolerance analysis of SAIR6 long non-coding RNA in Tamarix hispida[J]. J Beijing For Univ, 2021, 43(3): 36-43. | |
| [50] | 许欣. 刚毛柽柳盐胁迫响应长链非编码RNA的鉴定及耐盐功能分析[D]. 哈尔滨: 东北林业大学, 2021. |
| Xu X. Identification and functional analysis of long non-coding RNA on salt tolerance of Tamarix hispida[D]. Harbin: Northeast Forestry University, 2021. | |
| [51] | 赵雷. 拟南芥长链非编码RNA调控高温胁迫响应的机理研究[D]. 泰安: 山东农业大学, 2017. |
| Zhao L. Functional analysis of long nocoding RNA HAL6 for heat stress tolerance in Arabidopsis[D]. Tai'an: Shandong Agricultural University, 2017. | |
| [52] |
Wunderlich M, Gross-Hardt R, Schöffl F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA[J]. Plant Mol Biol, 2014, 85(6): 541-550.
doi: 10.1007/s11103-014-0202-0 pmid: 24874772 |
| [53] |
Kindgren P, Ard R, Ivanov M, et al. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation[J]. Nat Commun, 2018, 9(1): 4561.
doi: 10.1038/s41467-018-07010-6 pmid: 30385760 |
| [54] | 蔡晶晶. 长链非编码RNA13853调控拟南芥干旱胁迫应答机制研究[D]. 南昌: 南昌大学, 2020. |
| Cai JJ. Mechanism of Arabidopsis thaliana drought response regulated by a long noncoding RNA 13853[D]. Nanchang: Nanchang University, 2020. | |
| [55] | 毋若楠, 王红, 杨成成, 等. 拟南芥lncRNA-At5NC056820过表达载体构建及其转基因植株的抗旱性研究[J]. 西北植物学报, 2017, 37(10): 1904-1909. |
| Wu RN, Wang H, Yang CC, et al. Construction of lncRNA-At5NC056820 overexpression vector in Arabidopsis thaliana and study on drought resistance of transgenic plants[J]. Acta Bot Boreali Occidentalia Sin, 2017, 37(10): 1904-1909. | |
| [56] |
Qin T, Zhao HY, Cui P, et al. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance[J]. Plant Physiol, 2017, 175(3): 1321-1336.
doi: 10.1104/pp.17.00574 pmid: 28887353 |
| [57] |
Sun YQ, Hao PB, Lv XM, et al. A long non-coding apple RNA, MSTRG. 85814. 11, acts as a transcriptional enhancer of SAUR32 and contributes to the Fe-deficiency response[J]. Plant J, 2020, 103(1): 53-67.
doi: 10.1111/tpj.14706 URL |
| [58] |
Wu J, Okada T, Fukushima T, et al. A novel hypoxic stress-responsive long non-coding RNA transcribed by RNA polymerase III in Arabidopsis[J]. RNA Biol, 2012, 9(3): 302-313.
doi: 10.4161/rna.19101 URL |
| [59] | Li S, Nayar S, Jia HY, et al. The Arabidopsis hypoxia inducible AtR8 long non-coding RNA also contributes to plant defense and root elongation coordinating with WRKY genes under low levels of Sal Icylic acid[J]. Noncoding RNA, 2020, 6(1): 8. |
| [60] | 王亚丽. 玉米赤霉素响应长链非编码RNA发掘及lncRNA GARR2的功能研究[D]. 扬州: 扬州大学, 2021. |
| Wang YL. Identification of gibberellin-responsive long non-coding RNA and functional analysis of lncRNA gibberellin responsive lncRNA GARR2[D]. Yangzhou: Yangzhou University, 2021. | |
| [61] |
Wang D, Qu ZP, Yang L, et al. Transposable elements(TEs)contribute to stress-related long intergenic noncoding RNAs in plants[J]. Plant J, 2017, 90(1): 133-146.
doi: 10.1111/tpj.13481 URL |
| [62] |
Ariel F, Jegu T, Latrasse D, et al. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop[J]. Mol Cell, 2014, 55(3): 383-396.
doi: 10.1016/j.molcel.2014.06.011 pmid: 25018019 |
| [63] |
Cui J, Luan YS, Jiang N, et al. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin[J]. Plant J, 2017, 89(3): 577-589.
doi: 10.1111/tpj.13408 URL |
| [64] |
Jiang N, Cui J, Shi YS, et al. Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction[J]. Hortic Res, 2019, 6:28.
doi: 10.1038/s41438-018-0096-0 URL |
| [65] |
Cui J, Jiang N, Meng J, et al. LncRNA33732-respiratory burst oxidase module associated with WRKY1 in tomato- Phytophthora infestans interactions[J]. Plant J, 2019, 97(5): 933-946.
doi: 10.1111/tpj.14173 URL |
| [66] |
Hou XX, Cui J, Liu WW, et al. LncRNA39026 enhances tomato resistance to Phytophthora infestans by decoying miR168a and inducing PR gene expression[J]. Phytopathology, 2020, 110(4): 873-880.
doi: 10.1094/PHYTO-12-19-0445-R URL |
| [67] |
Wang JY, Yang YW, Jin LM, et al. Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection[J]. BMC Plant Biol, 2018, 18(1): 104.
doi: 10.1186/s12870-018-1332-3 pmid: 29866032 |
| [68] | Yang YW, Liu TL, Shen DY, et al. Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms[J]. PLoS Pathog, 2019, 15(1): e1007534. |
| [69] |
Yu Y, Zhou YF, Feng YZ, et al. Transcriptional landscape of pathogen-responsive lncRNAs in rice unveils the role of ALEX1 in jasmonate pathway and disease resistance[J]. Plant Biotechnol J, 2020, 18(3): 679-690.
doi: 10.1111/pbi.13234 URL |
| [70] |
Zhu QH, Stephen S, Taylor J, et al. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana[J]. New Phytol, 2014, 201(2): 574-584.
doi: 10.1111/nph.12537 URL |
| [71] |
Seo JS, Sun HX, Park BS, et al. ELF18-induced long-noncoding RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis[J]. Plant Cell, 2017, 29(5): 1024-1038.
doi: 10.1105/tpc.16.00886 URL |
| [72] |
Gao RM, Liu P, Irwanto N, et al. Upregulation of LINC-AP2 is negatively correlated with AP2 gene expression with Turnip crinkle virus infection in Arabidopsis thaliana[J]. Plant Cell Rep, 2016, 35(11): 2257-2267.
doi: 10.1007/s00299-016-2032-9 URL |
| [73] |
Zhang L, Wang MJ, Li NN, et al. Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton[J]. Plant Biotechnol J, 2018, 16(6): 1172-1185.
doi: 10.1111/pbi.12861 pmid: 29149461 |
| [1] | ZHAN Yan, ZHOU Li-bin, JIN Wen-jie, DU Yan, YU Li-xia, QU Ying, MA Yong-gui, LIU Rui-yuan. Research Progress in Plant Leaf Color Mutation Induced by Radiation [J]. Biotechnology Bulletin, 2023, 39(8): 106-113. |
| [2] | WANG Bao-bao, WANG Hai-yang. Molecular Design of Ideal Plant Architecture for High-density Tolerance of Maize Plant [J]. Biotechnology Bulletin, 2023, 39(8): 11-30. |
| [3] | JIANG Run-hai, JIANG Ran-ran, ZHU Cheng-qiang, HOU Xiu-li. Research Progress in Mechanisms of Microbial-enhanced Phytoremediation for Lead-contaminated Soil [J]. Biotechnology Bulletin, 2023, 39(8): 114-125. |
| [4] | WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq [J]. Biotechnology Bulletin, 2023, 39(8): 173-184. |
| [5] | LIU Bao-cai, CHEN Jing-ying, ZHANG Wu-jun, HUANG Ying-zhen, ZHAO Yun-qing, LIU Jian-chao, WEI Zhi-cheng. Characteristics Analysis of Seed Microrhizome Gene Expression of Polygonatum cyrtonema [J]. Biotechnology Bulletin, 2023, 39(8): 220-233. |
| [6] | SHI Jia-xin, LIU Kai, ZHU Jin-jie, QI Xian-tao, XIE Chuan-xiao, LIU Chang-lin. Gene Editing Reshaping Maize Plant Type for Increasing Hybrid Yield [J]. Biotechnology Bulletin, 2023, 39(8): 62-69. |
| [7] | ZHANG Yong, XU Tian-jun, LYU Tian-fang, XING Jin-feng, LIU Hong-wei, CAI Wan-tao, LIU Yue-e, ZHAO Jiu-ran, WANG Rong-huan. Effects of Planting Density on the Stem Quality and Root Phenotypic Characters of Summer Sowing Maize [J]. Biotechnology Bulletin, 2023, 39(8): 70-79. |
| [8] | YAO Sha-sha, WANG Jing-jing, WANG Jun-jie, LIANG Wei-hong. Molecular Mechanisms of Rice Grain Size Regulation Related to Plant Hormone Signaling Pathways [J]. Biotechnology Bulletin, 2023, 39(8): 80-90. |
| [9] | ZHANG Man, ZHANG Ye-zhuo, HE Qi-zou-hong, E Yi-lan, LI Ye. Advances in Plant Cell Wall Structure and Imaging Technology [J]. Biotechnology Bulletin, 2023, 39(7): 113-122. |
| [10] | HU Hai-lin, XU Li, LI Xiao-xu, WANG Chen-can, MEI Man, DING Wen-jing, ZHAO Yuan-yuan. Advances in the Regulation of Plant Growth, Development and Stress Physiology by Small Peptide Hormones [J]. Biotechnology Bulletin, 2023, 39(7): 13-25. |
| [11] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [12] | XU Jian-xia, DING Yan-qing, FENG Zhou, CAO Ning, CHENG Bin, GAO Xu, ZOU Gui-hua, ZHANG Li-yi. QTL Mapping of Sorghum Plant Height and Internode Numbers Based on Super-GBS Technique [J]. Biotechnology Bulletin, 2023, 39(7): 185-194. |
| [13] | LI Ying, YUE Xiang-hua. Application of DNA Methylation in Interpreting Natural Variation in Moso Bamboo [J]. Biotechnology Bulletin, 2023, 39(7): 48-55. |
| [14] | LI Yu-ling, MAO Xin, ZHANG Yuan-shuai, DONG Yuan-fu, LIU Cui-lan, DUAN Chun-hua, MAO Xiu-hong. Applications and Perspectives of Radiation Mutagenesis in Woody Plant Breeding [J]. Biotechnology Bulletin, 2023, 39(6): 12-30. |
| [15] | YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum [J]. Biotechnology Bulletin, 2023, 39(6): 259-273. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||