








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (10): 197-208.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0364
Previous Articles Next Articles
WANG Bing1( ), ZHAO Hui-na1, YU Jing1, CHEN Jie1, LUO Mei2, LEI Bo1(
), ZHAO Hui-na1, YU Jing1, CHEN Jie1, LUO Mei2, LEI Bo1( )
)
Received:2023-04-19
Online:2023-10-26
Published:2023-11-28
Contact:
LEI Bo
E-mail:vipwb1519599@163.com;leibo_1981@163.com
WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System[J]. Biotechnology Bulletin, 2023, 39(10): 197-208.
| 引物Primer | 序列Sequence(5'-3') | 作用Purpose |
|---|---|---|
| C16NtREV-T1F | ATTGGCTCGATATTCGACAGAATAGGG | 靶点接头引物 NtREV target construction |
| C16NtREV-T1R | AAACCCCTATTCTGTCGAATATCGAGC | |
| C15NtREV-T1F | ATTGACAGTAGTGGAAAGTATGTCCGG | |
| C15NtREV-T1R | AAACCCGGACATACTTTCCACTACTGT | |
| 1300-gRNA-F | CCAGTCACGACGTTGTAAAAC | SgRNA表达盒检测 Detection of SgRNA expression kit |
| 1300-gRNA-R | CAATGAATTTCCCATCGTCGAG | |
| Hyg-F | CGATTGCGTCGCATCGACC | 潮霉素抗性基因检测 Detection of hygromycin resistance gene |
| Hyg-R | TTCTACAACCGGTCGCGGAG | |
| C16NtREV-CRISPR test-F | TTTTGGTTTGGGATTTTGAGG | 检测转基因突变 Detection of transgenic mutations |
| C16NtREV-CRISPR test-R | CATTGTTCAATTTGGATTACTCC | |
| C16NtREV-CRISPR test-R | GGAGCAATCAAAATTGAACAGCG | |
| C15NtREV-CRISPR test-F | CTATGGTTGCACAGCAGCACA | |
| C15NtREV-CRISPR test-R | ACTTCATCCATCACTGATCTAAC | |
| C15NtREV-CRISPR test-R | ACTAGAAGCACAGTGATATATTC |
Table 1 Sequences and purpose of primers in this study
| 引物Primer | 序列Sequence(5'-3') | 作用Purpose |
|---|---|---|
| C16NtREV-T1F | ATTGGCTCGATATTCGACAGAATAGGG | 靶点接头引物 NtREV target construction |
| C16NtREV-T1R | AAACCCCTATTCTGTCGAATATCGAGC | |
| C15NtREV-T1F | ATTGACAGTAGTGGAAAGTATGTCCGG | |
| C15NtREV-T1R | AAACCCGGACATACTTTCCACTACTGT | |
| 1300-gRNA-F | CCAGTCACGACGTTGTAAAAC | SgRNA表达盒检测 Detection of SgRNA expression kit |
| 1300-gRNA-R | CAATGAATTTCCCATCGTCGAG | |
| Hyg-F | CGATTGCGTCGCATCGACC | 潮霉素抗性基因检测 Detection of hygromycin resistance gene |
| Hyg-R | TTCTACAACCGGTCGCGGAG | |
| C16NtREV-CRISPR test-F | TTTTGGTTTGGGATTTTGAGG | 检测转基因突变 Detection of transgenic mutations |
| C16NtREV-CRISPR test-R | CATTGTTCAATTTGGATTACTCC | |
| C16NtREV-CRISPR test-R | GGAGCAATCAAAATTGAACAGCG | |
| C15NtREV-CRISPR test-F | CTATGGTTGCACAGCAGCACA | |
| C15NtREV-CRISPR test-R | ACTTCATCCATCACTGATCTAAC | |
| C15NtREV-CRISPR test-R | ACTAGAAGCACAGTGATATATTC |
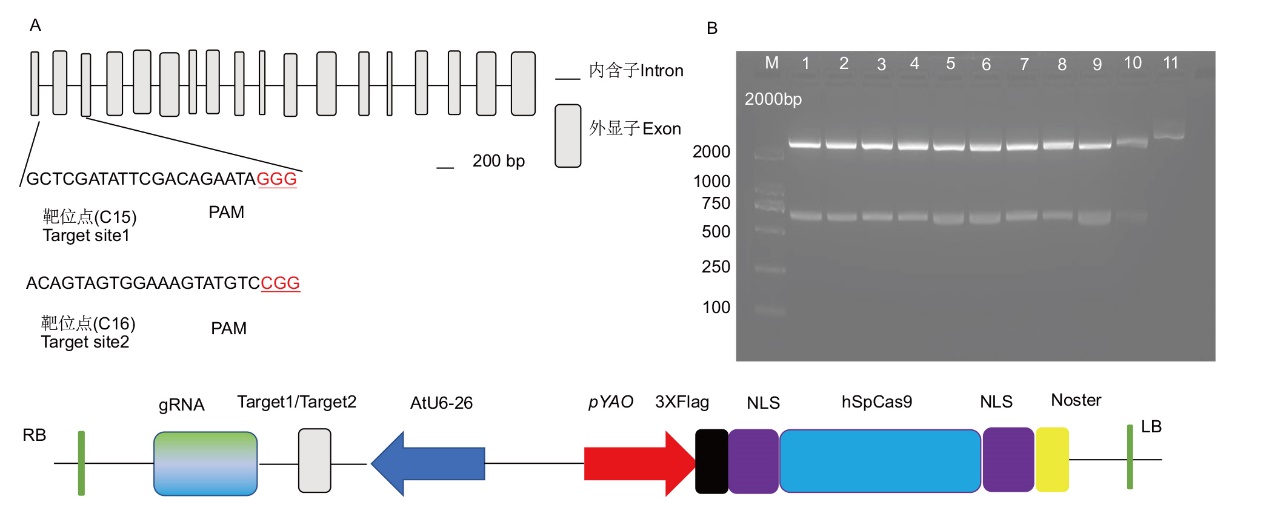
Fig. 2 Schematic structure and target site selection of NtREV and vector construction A: Schematic diagram of REV gene exons and introns, as well as the locations of the target editing sequences. B: Nucleic acid electrophoresis of double enzyme digestion of REV gene the target editing sequences
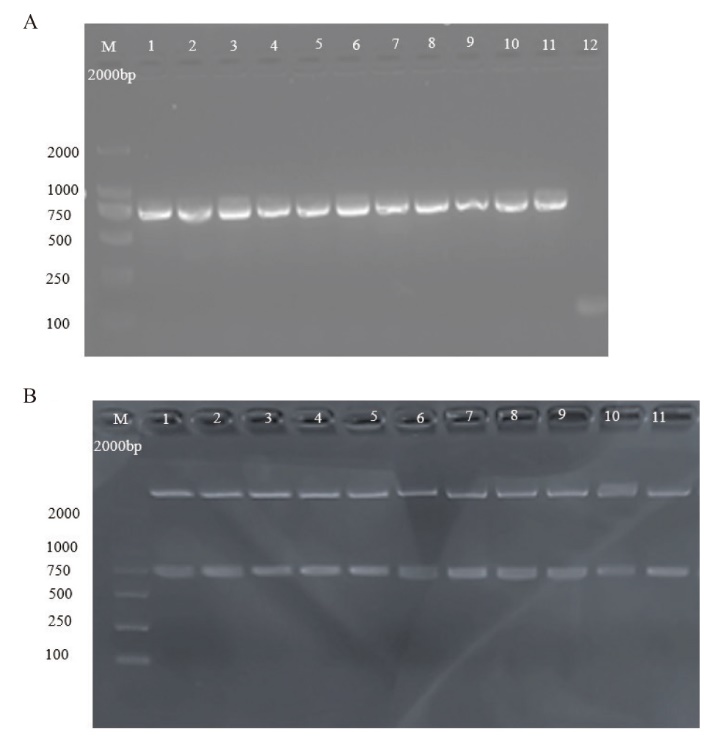
Fig. 3 Construction of the plant binary expression vector pCAMBIA1300-pYAO-Cas9-NtREV A: Nucleic acid electrophoresis of PCR amplification of pCAMBIA1300-pYAO-Cas9-NtREV. B: Nucleic acid electrophoresis of double enzyme digestion of pCAMBIA1300-pYAO-Cas9-NtREV
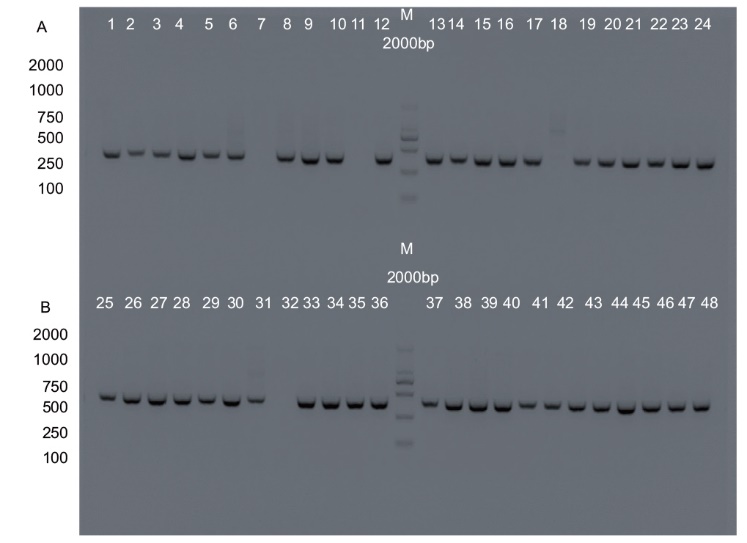
Fig. 4 Nucleic acid electrophoresis for the detection of transgenic plants A: Nucleic acid electrophoresis for detecting plant editing of the REV gene at target C15. B: Nucleic acid electrophoresis for detecting plant editing of the REV gene at target C16

Fig. 5 Mutation form analysis of NtREV edited by the CRISPR/Cas9 A: Mutation of REV target C16 sequence; B: mutation of REV target C15 sequence. Underscores indicate PAM sequences, blue words indicate replacement bases, red words indicate insertion bases, and - indicates missing bases

Fig. 6 Leaf morphological phenotypes of mutant and wild-type materials A-F: Leaf development morphology of the double-copy homologous mutants, including funnel-shaped leaves, lotus-shaped leaves and other morphologies; G-H: leaf development morphology of the single-copy homologous mutants; I: wild type growth state in the corresponding period; bar=1 cm
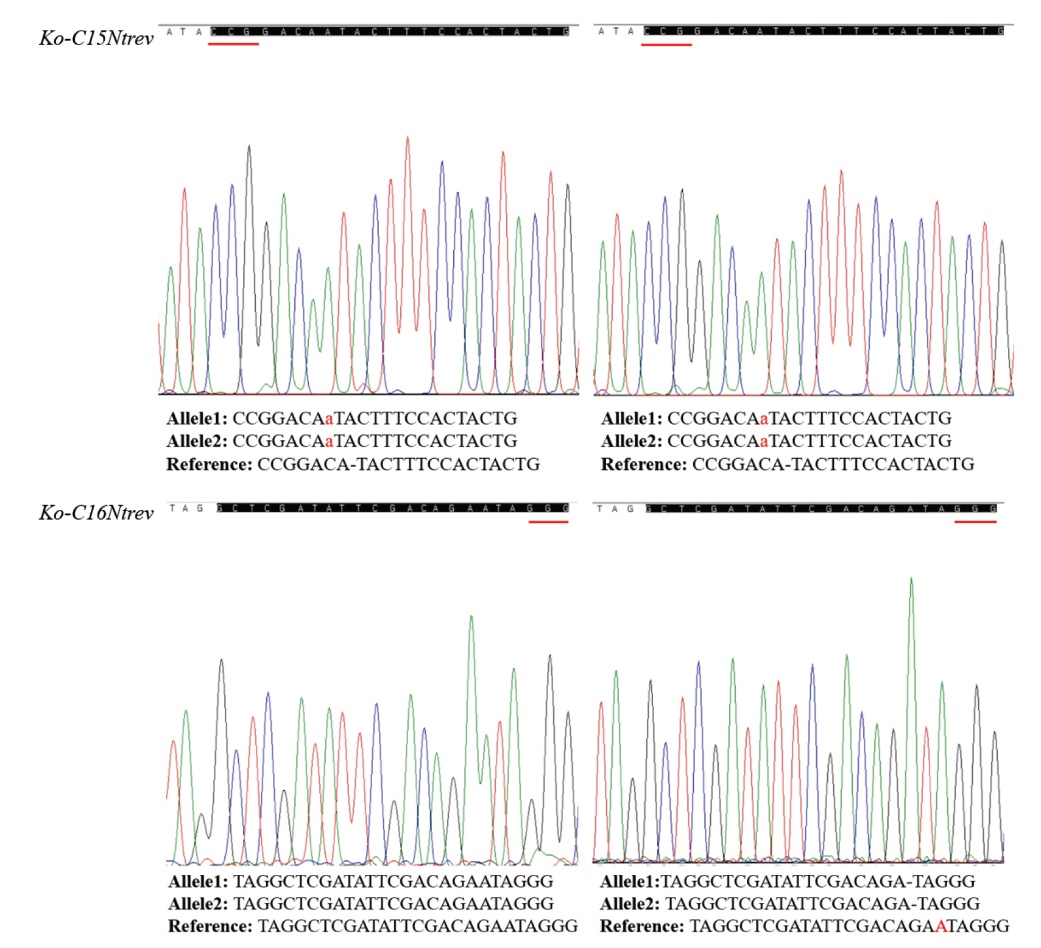
Fig. 7 Editing forms of Ko-C15Ntrev and Ko-C16Ntrev mutants in tobacco The double-copy homologous mutants of Ko-C15NtREV; Ko-C16NtREV editing form is the single-copy homologous mutants
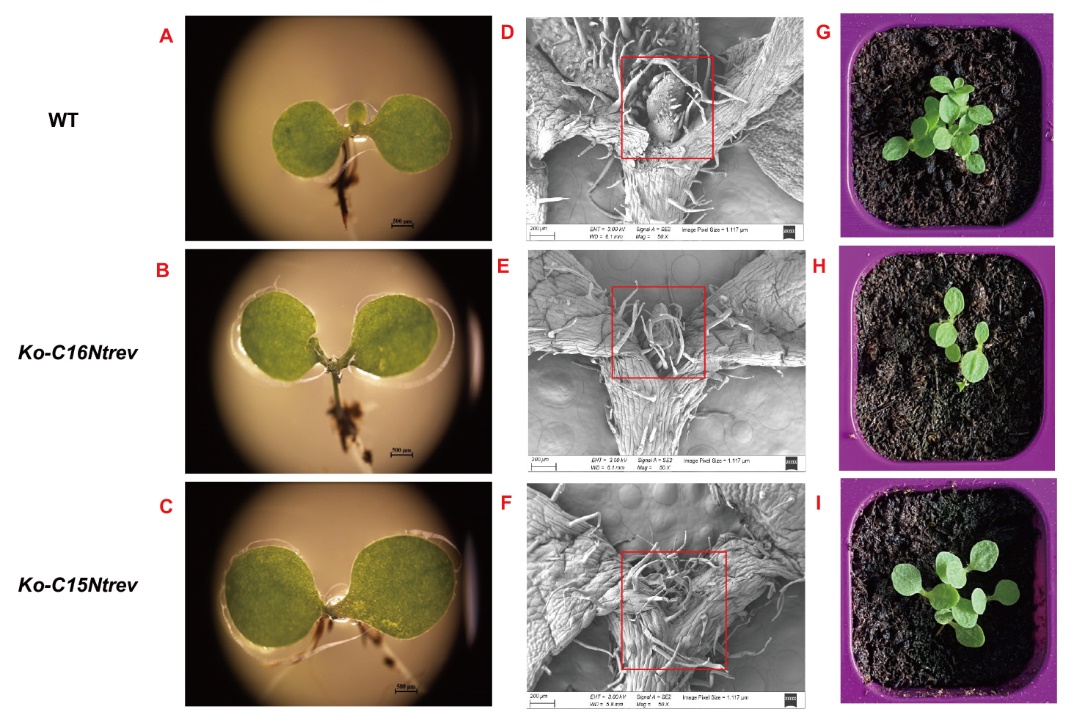
Fig. 8 Observations of apical buds of mutant and wild-type ones A, B, C: Apical bud phenotypes of tobacco seedling wild-type, Ko-C16Ntrev and Ko-C15Ntrev mutant under stereomicroscope, respectively; D, E, F: apical meristem phenotypes of tobacco seedling wild-type,Ko-C16Ntrev and Ko-C15Ntrev mutant under scanning electron microscopy, respectively; G, H, I: tobacco seedling wild-type, Ko-C16Ntrev and Ko-C15Ntrev mutant corresponding to the above phenotypes, respectively

Fig. 9 Phenotypes of plant height, leaf numbers and fresh weight of C16Ntrev mutants A, B, C: Statistical analysis of plant height, leaf numbers and fresh weight content of axillary buds of WT and Ko-C16Ntrev mutants. D and E: The phenotype of lateral buds of WT and Ko-C16Ntrev mutants, respectively. Bar= 5 cm. The data represent mean ± SD, n ≥ 10; ** indicates significance P<0.01
| [1] |
Teichmann T, Muhr M. Shaping plant architecture[J]. Front Plant Sci, 2015, 6: 233.
doi: 10.3389/fpls.2015.00233 pmid: 25914710 |
| [2] |
王兵, 赵会纳, 余婧, 等. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1112 |
|
Wang B, Zhao HN, Yu J, et al. Research progress in the regulation of plant branch development[J]. Biotechnol Bull, 2023, 39(5): 14-22.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1112 |
|
| [3] |
Luo M, Gilbert B, Ayliffe M. Applications of CRISPR/Cas9 technology for targeted mutagenesis, gene replacement and stacking of genes in higher plants[J]. Plant Cell Rep, 2016, 35(7): 1439-1450.
doi: 10.1007/s00299-016-1989-8 pmid: 27146973 |
| [4] | 刘耀光, 李构思, 张雅玲, 等. CRISPR/Cas植物基因组编辑技术研究进展[J]. 华南农业大学学报, 2019, 40(5): 38-49. |
| Liu YG, Li GS, Zhang YL, et al. Current advances on CRISPR/Cas genome editing technologies in plants[J]. J South China Agric Univ, 2019, 40(5): 38-49. | |
| [5] | 杨雪, 孙雅佩, 王政博, 等. CRISPR/Cas9介导靶向敲除拟南芥REVOLUTA基因突变体的鉴定[J]. 分子植物育种, 2021, 19(3): 867-873. |
| Yang X, Sun YP, Wang ZB, et al. Identification of knockout of REVOLUTA mutant caused by CRISPR/Cas9 in Arabidopsis[J]. Mol Plant Breed, 2021, 19(3): 867-873. | |
| [6] | Amoo O, 胡利民, 翟云孤, 等. 利用基因编辑技术研究BRANCHED1参与油菜分枝过程的调控[J]. 生物技术通报, 2022, 38(4): 97-105. |
| Amoo O, Hu LM, Zhai YG, et al. Regulation of shoot branching by BRANCHED1 in Brassica napus based on gene editing technology[J]. Biotechnol Bull, 2022, 38(4): 97-105. | |
| [7] |
Prigge MJ, Otsuga D, Alonso JM, et al. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development[J]. Plant Cell, 2005, 17(1): 61-76.
doi: 10.1105/tpc.104.026161 URL |
| [8] |
Zhong RQ, Ye ZH. Regulation of HD-ZIP III genes by microRNA 165[J]. Plant Signal Behav, 2007, 2(5): 351-353.
doi: 10.4161/psb.2.5.4119 URL |
| [9] |
Kim J, Jung JH, Reyes JL, et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems[J]. Plant J, 2005, 42(1): 84-94.
doi: 10.1111/tpj.2005.42.issue-1 URL |
| [10] | 杜玉梁, 陈叶, 刘彩虹, 等. 植物维管组织形态建成的分子调控机制[J]. 北京林业大学学报, 2014, 36(3): 142-150. |
| Du YL, Chen Y, Liu CH, et al. Molecular regulation mechanism of vascular pattern formation in plant[J]. J Beijing For Univ, 2014, 36(3): 142-150. | |
| [11] |
Talbert PB, Adler HT, Parks DW, et al. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana[J]. Development, 1995, 121(9): 2723-2735.
doi: 10.1242/dev.121.9.2723 pmid: 7555701 |
| [12] |
Otsuga D, DeGuzman B, Prigge MJ, et al. REVOLUTA regulates meristem initiation at lateral positions[J]. Plant J, 2001, 25(2): 223-236.
doi: 10.1046/j.1365-313x.2001.00959.x pmid: 11169198 |
| [13] |
Williams L, Grigg SP, Xie MT, et al. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes[J]. Development, 2005, 132(16): 3657-3668.
doi: 10.1242/dev.01942 pmid: 16033795 |
| [14] |
Zhong RQ, Ye ZH. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels[J]. Plant Cell Physiol, 2004, 45(4): 369-385.
doi: 10.1093/pcp/pch051 URL |
| [15] |
Hawker NP, Bowman JL. Roles for Class III HD-Zip and KANADI genes in Arabidopsis root development[J]. Plant Physiol, 2004, 135(4): 2261-2270.
doi: 10.1104/pp.104.040196 URL |
| [16] |
Shi BH, Zhang C, Tian CH, et al. Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis[J]. PLoS Genet, 2016, 12(7): e1006168.
doi: 10.1371/journal.pgen.1006168 URL |
| [17] | Zhang C, Wang J, Wenkel S, et al. Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators[J]. Development, 2018, 145(24): dev158352. |
| [18] |
Zhang C, Fan LS, Le BH, et al. Regulation of ARGONAUTE10 expression enables temporal and spatial precision in axillary meristem initiation in Arabidopsis[J]. Dev Cell, 2020, 55(5): 603-616.e5.
doi: 10.1016/j.devcel.2020.10.019 pmid: 33232670 |
| [19] | 陈浣, 吕婧, 孙玉合. 普通烟草HD-Zip III家族全基因组鉴定和表达分析[J]. 基因组学与应用生物学, 2017, 36(8): 3034-3041. |
| Chen H, Lyu J, Sun YH. Genome-wide identification and expression analysis of the HD-zip III gene family in Nicotiana tobacum[J]. Genom Appl Biol, 2017, 36(8): 3034-3041. | |
| [20] |
Ma XL, Zhang QY, Zhu QL, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants[J]. Mol Plant, 2015, 8(8): 1274-1284.
doi: 10.1016/j.molp.2015.04.007 pmid: 25917172 |
| [21] | 王兵, 程子义, 张蕾, 等. 过表达毛白杨线粒体APX基因烟草提高抗逆能力的研究[J]. 北京林业大学学报, 2020, 42(7): 33-39. |
| Wang B, Cheng ZY, Zhang L, et al. Tobacco overexpression Populus tomentosa mitochondria ascorbate peroxidase improving stress resistance[J]. J Beijing For Univ, 2020, 42(7): 33-39. | |
| [22] | 王兵.毛白杨天冬氨酸蛋白酶(PtoAED3)调控次生木质部发育机制研究[D]. 北京: 北京林业大学, 2020. |
| Wang B.Study on the mechanism of secondary xylem development regulated by aspartic protease(PtoAED3) of Populus tomentosa[D]. Beijing: Beijing Forestry University, 2020. | |
| [23] |
Zhang T, You J, Zhang Y, et al. LF1 regulates the lateral organs polarity development in rice[J]. New Phytol, 2021, 231(3): 1265-1277.
doi: 10.1111/nph.17220 pmid: 33469925 |
| [24] |
张晨, 雷展, 李凯, 等. CRISPR/Cas9系统中的脱靶效应及检测技术研究进展[J]. 生物技术通报, 2020, 36(3): 78-87.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0871 |
|
Zhang C, Lei Z, Li K, et al. Research progress on off-target effects and detection techniques in CRISPR/Cas9 systems[J]. Biotechnol Bull, 2020, 36(3): 78-87.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0871 |
|
| [25] |
Mukherjee K, Bürglin TR. MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins[J]. Plant Physiol, 2006, 140(4): 1142-1150.
doi: 10.1104/pp.105.073833 pmid: 16607028 |
| [26] |
Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains[J]. Structure, 2009, 17(10): 1282-1294.
doi: 10.1016/j.str.2009.08.011 pmid: 19836329 |
| [27] |
McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim proteins in environmental adaptation[J]. Annu Rev Physiol, 2010, 72: 625-645.
doi: 10.1146/annurev-physiol-021909-135922 pmid: 20148691 |
| [28] |
Park SY, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins[J]. Science, 2009, 324(5930): 1068-1071.
doi: 10.1126/science.1173041 URL |
| [29] |
Brandt R, Salla-Martret M, Bou-Torrent J, et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses[J]. Plant J, 2012, 72(1): 31-42.
doi: 10.1111/tpj.2012.72.issue-1 URL |
| [1] | WANG Bao-bao, WANG Hai-yang. Molecular Design of Ideal Plant Architecture for High-density Tolerance of Maize Plant [J]. Biotechnology Bulletin, 2023, 39(8): 11-30. |
| [2] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [3] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [4] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [5] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [6] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [7] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [8] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [9] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [10] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [11] | Olalekan Amoo, HU Li-min, ZHAI Yun-gu, FAN Chu-chuan, ZHOU Yong-ming. Regulation of Shoot Branching by BRANCHED1 in Brassica napus Based on Gene Editing Technology [J]. Biotechnology Bulletin, 2022, 38(4): 97-105. |
| [12] | DING Ya-qun, DING Ning, XIE Shen-min, HUANG Meng-na, ZHANG Yu, ZHANG Qin, JIANG Li. Construction of Vps28 Knock-out Mice and Model Study of the Impact on Lactation and Immune Traits [J]. Biotechnology Bulletin, 2022, 38(3): 164-172. |
| [13] | YAN Jiong, FENG Chen-yi, GAO Xue-kun, XU Xiang, YANG Jia-min, CHEN Zhao-yang. Construction of Homozygous Plin1-knockout Mouse Model and Phenotype Analysis Based on CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2022, 38(3): 173-180. |
| [14] | ZHONG Jing, SUN Ling-ling, ZHANG Shu, MENG Yuan, ZHI Yi-fei, TU Li-qing, XU Tian-peng, PU Li-ping, LU Yang-qing. Effect of Knocking Out the Mda5 Gene by CRISPR/Cas9 Technology on the Replication of Newcastle Disease and Infectious Bursal Virus [J]. Biotechnology Bulletin, 2022, 38(11): 90-96. |
| [15] | ZONG Mei, HAN Shuo, GUO Ning, DUAN Meng-meng, LIU Fan, WANG Gui-xiang. Production of Marker-free Mutants of Brassica campestris Mediated by CRISPR/Cas9 Through Vacuum Infiltration [J]. Biotechnology Bulletin, 2022, 38(10): 159-163. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||