








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (11): 205-216.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0722
Previous Articles Next Articles
TANG Rui-qi1( ), ZHAO Xin-qing2, ZHU Du1, WANG Ya1(
), ZHAO Xin-qing2, ZHU Du1, WANG Ya1( )
)
Received:2023-07-28
Online:2023-11-26
Published:2023-12-20
Contact:
WANG Ya
E-mail:rq_tang@jxstnu.edu.cn;wangya@jxstnu.edu.cn
TANG Rui-qi, ZHAO Xin-qing, ZHU Du, WANG Ya. Stress Tolerance of Escherichia coli to Inhibitors in Lignocellulosic Hydrolysates[J]. Biotechnology Bulletin, 2023, 39(11): 205-216.
| 基因 Gene | 敲除/过表达 Deletion/Overexpression | 描述 Description | 抑制物 Inhibitor | 参考文献 Reference |
|---|---|---|---|---|
| pgi, encoding glucose-6-phosphate isomerase | Deletion | Shunt glucose to pentose phosphate pathway to increase NADPH production | Furfural, HMF | [ |
| pntAB, encoding pyridine nucleotide transhydrogenase | Overexpression | Convert NADP+ to NADPH using PntAB to increase NADPH regeneration | Furfural, HMF | [ |
| yqhD, encoding aldehyde reductase | Deletion | Delete NADPH-dependent YqhD to reduce NADPH consumption | Furfural, HMF | [ |
| yqhC, encoding transcriptional activator | Deletion | Delete YqhC to downregulate yqhDexpression, reducing NADPH consumption | Furfural | [ |
| fucO, encoding propanediol oxidoreductase | Overexpression | Convert furfural using NADH-dependent FucO to reduce NADPH consumption | Furfural | [ |
| yghA, encoding oxidoreductase | Overexpression | Convert furfural using NADH-dependent YghA to reduce NADPH consumption | Furfural, HMF | [ |
| pncB and nadE, encoding NAD salvage pathway enzymes | Overexpression | Increase NAD(P)H level through the nicotine amide salvage pathway | Furfural | [ |
| Heterologous xylBand BaBAD, encoding benzyl alcohol dehydrogenases | Overexpression | Convert furfural using NADH-dependent XylB and BaBAD to reduce NADPH consumption | Furfural | [ |
| thyA, encoding thymidylate synthase | Overexpression | Overexpress ThyA to increase dTMP level for DNA repair | Furfural | [ |
| potE and puuP, encoding polyamine transporters | Overexpression | Increase cytoplasmic polyamine level to maintain DNA synthesis | Furfural | [ |
| lpcA, encoding D-sedoheptulose-7-phosphate isomerase | Overexpression | Overexpress LpcA to increase formation of lipopolysaccharides and NADPH | Furfural | [ |
| pssA, encoding phosphatidylserine synthase | Overexpression | Increase phosphatidylethanolamine content to increase membrane integrity | Furfural, HMF | [ |
| mdtJI, encoding multidrug resistance efflux pump | Overexpression | Export furfural by efflux pump MdtJI | Furfural, HMF | [ |
| groESL, encoding chaperonin | Overexpression | Maintain proper folding of proteins | Furfural | [ |
Table 1 Gene targets for improving the tolerance of E. coli to furan inhibitors
| 基因 Gene | 敲除/过表达 Deletion/Overexpression | 描述 Description | 抑制物 Inhibitor | 参考文献 Reference |
|---|---|---|---|---|
| pgi, encoding glucose-6-phosphate isomerase | Deletion | Shunt glucose to pentose phosphate pathway to increase NADPH production | Furfural, HMF | [ |
| pntAB, encoding pyridine nucleotide transhydrogenase | Overexpression | Convert NADP+ to NADPH using PntAB to increase NADPH regeneration | Furfural, HMF | [ |
| yqhD, encoding aldehyde reductase | Deletion | Delete NADPH-dependent YqhD to reduce NADPH consumption | Furfural, HMF | [ |
| yqhC, encoding transcriptional activator | Deletion | Delete YqhC to downregulate yqhDexpression, reducing NADPH consumption | Furfural | [ |
| fucO, encoding propanediol oxidoreductase | Overexpression | Convert furfural using NADH-dependent FucO to reduce NADPH consumption | Furfural | [ |
| yghA, encoding oxidoreductase | Overexpression | Convert furfural using NADH-dependent YghA to reduce NADPH consumption | Furfural, HMF | [ |
| pncB and nadE, encoding NAD salvage pathway enzymes | Overexpression | Increase NAD(P)H level through the nicotine amide salvage pathway | Furfural | [ |
| Heterologous xylBand BaBAD, encoding benzyl alcohol dehydrogenases | Overexpression | Convert furfural using NADH-dependent XylB and BaBAD to reduce NADPH consumption | Furfural | [ |
| thyA, encoding thymidylate synthase | Overexpression | Overexpress ThyA to increase dTMP level for DNA repair | Furfural | [ |
| potE and puuP, encoding polyamine transporters | Overexpression | Increase cytoplasmic polyamine level to maintain DNA synthesis | Furfural | [ |
| lpcA, encoding D-sedoheptulose-7-phosphate isomerase | Overexpression | Overexpress LpcA to increase formation of lipopolysaccharides and NADPH | Furfural | [ |
| pssA, encoding phosphatidylserine synthase | Overexpression | Increase phosphatidylethanolamine content to increase membrane integrity | Furfural, HMF | [ |
| mdtJI, encoding multidrug resistance efflux pump | Overexpression | Export furfural by efflux pump MdtJI | Furfural, HMF | [ |
| groESL, encoding chaperonin | Overexpression | Maintain proper folding of proteins | Furfural | [ |
| 基因 Gene | 描述 Description | 胁迫 Stress | 参考文献 Reference |
|---|---|---|---|
| dsrA and hfq, encoding small noncoding RNA and chaperone | DsrA increases rpoS mRNA stability and activate RpoS translation, Hfq promotes DsrA annealing to the rpoS5' untranscribed region(UTR) | Low pH | [ |
| Heterologous cfaS, encoding cyclopropane fatty acid synthase | Decrease membrane permeability and fluidity | Low pH | [ |
| Heterologous cbpA, encoding chaperone | CbpA plays a role in protein and DNA repair | Acetate | [ |
| gadE, encoding transcriptional activator | GadE activates acid resistance system | Low pH | [ |
| hdeB, encoding periplasmic acid stress chaperone | HdeB prevents periplasmic proteins aggregation at low pH | Low pH | [ |
| sodB and katE, encoding superoxide dismutase and catalase | SodB and KatE are ROS scavengers | Low pH | [ |
Table 2 Overexpression targets for improving the tolerance of E. coli to acid
| 基因 Gene | 描述 Description | 胁迫 Stress | 参考文献 Reference |
|---|---|---|---|
| dsrA and hfq, encoding small noncoding RNA and chaperone | DsrA increases rpoS mRNA stability and activate RpoS translation, Hfq promotes DsrA annealing to the rpoS5' untranscribed region(UTR) | Low pH | [ |
| Heterologous cfaS, encoding cyclopropane fatty acid synthase | Decrease membrane permeability and fluidity | Low pH | [ |
| Heterologous cbpA, encoding chaperone | CbpA plays a role in protein and DNA repair | Acetate | [ |
| gadE, encoding transcriptional activator | GadE activates acid resistance system | Low pH | [ |
| hdeB, encoding periplasmic acid stress chaperone | HdeB prevents periplasmic proteins aggregation at low pH | Low pH | [ |
| sodB and katE, encoding superoxide dismutase and catalase | SodB and KatE are ROS scavengers | Low pH | [ |
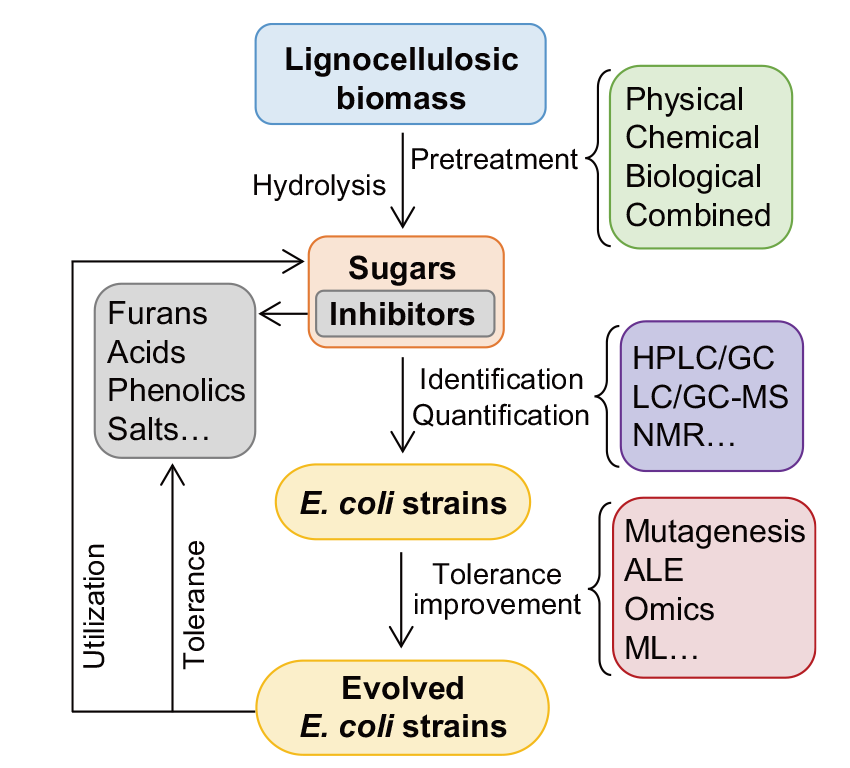
Fig. 3 Schematic diagram of the tolerance improvement of E. coli to inhibitors in lignocellulosic hydrolysates HPLC/GC indicate high-performance liquid chromatography/gas chromatography, LC-/GC-MS indicate liquid chromatography-/gas chromatography-mass spectrometer, NMR indicates nuclear magnetic resonance, ALE indicates adaptive laboratory evolution, ML indicates machine learning
| [1] |
Lu HD, Yadav V, Bilal M, et al. Bioprospecting microbial hosts to valorize lignocellulose biomass - Environmental perspectives and value-added bioproducts[J]. Chemosphere, 2022, 288(Pt 2): 132574.
doi: 10.1016/j.chemosphere.2021.132574 URL |
| [2] |
Reshmy R, Philip E, Madhavan A, et al. Lignocellulose in future biorefineries: strategies for cost-effective production of biomaterials and bioenergy[J]. Bioresour Technol, 2022, 344(Pt B): 126241.
doi: 10.1016/j.biortech.2021.126241 URL |
| [3] |
Zhai R, Hu JG, Jin MJ. Towards efficient enzymatic saccharification of pretreated lignocellulose: enzyme inhibition by lignin-derived phenolics and recent trends in mitigation strategies[J]. Biotechnol Adv, 2022, 61: 108044.
doi: 10.1016/j.biotechadv.2022.108044 URL |
| [4] |
Tan JY, Li Y, Tan X, et al. Advances in pretreatment of straw biomass for sugar production[J]. Front Chem, 2021, 9: 696030.
doi: 10.3389/fchem.2021.696030 URL |
| [5] |
Zhao CH, Zhang YP, Li Y. Production of fuels and chemicals from renewable resources using engineered Escherichia coli[J]. Biotechnol Adv, 2019, 37(7): 107402.
doi: 10.1016/j.biotechadv.2019.06.001 URL |
| [6] | Banerjee S, Pandit C, Gundupalli MP, et al. Life cycle assessment of revalorization of lignocellulose for the development of biorefineries[J]. Environ Dev Sustain, 2023. https://doi.org/10.1007/s10668-023-03360-4. |
| [7] |
Zhou M, Tian XJ. Development of different pretreatments and related technologies for efficient biomass conversion of lignocellulose[J]. Int J Biol Macromol, 2022, 202: 256-268.
doi: 10.1016/j.ijbiomac.2022.01.036 pmid: 35032493 |
| [8] |
Haldar D, Purkait MK. A review on the environment-friendly emerging techniques for pretreatment of lignocellulosic biomass: Mechanistic insight and advancements[J]. Chemosphere, 2021, 264(Pt 2): 128523.
doi: 10.1016/j.chemosphere.2020.128523 URL |
| [9] |
Shen XJ, Sun RC. Recent advances in lignocellulose prior-fractionation for biomaterials, biochemicals, and bioenergy[J]. Carbohydr Polym, 2021, 261: 117884.
doi: 10.1016/j.carbpol.2021.117884 URL |
| [10] |
Zhang R, Gao HR, Wang YT, et al. Challenges and perspectives of green-like lignocellulose pretreatments selectable for low-cost biofuels and high-value bioproduction[J]. Bioresour Technol, 2023, 369: 128315.
doi: 10.1016/j.biortech.2022.128315 URL |
| [11] |
van der Pol EC, Bakker RR, Baets P, et al. By-products resulting from lignocellulose pretreatment and their inhibitory effect on fermentations for(bio)chemicals and fuels[J]. Appl Microbiol Biotechnol, 2014, 98(23): 9579-9593.
doi: 10.1007/s00253-014-6158-9 URL |
| [12] |
Gallego-García M, Moreno AD, Manzanares P, et al. Recent advances on physical technologies for the pretreatment of food waste and lignocellulosic residues[J]. Bioresour Technol, 2023, 369: 128397.
doi: 10.1016/j.biortech.2022.128397 URL |
| [13] |
Shukla A, Kumar D, Girdhar M, et al. Strategies of pretreatment of feedstocks for optimized bioethanol production: distinct and integrated approaches[J]. Biotechnol Biofuels Bioprod, 2023, 16(1): 44.
doi: 10.1186/s13068-023-02295-2 |
| [14] | Wu ZY, Peng K, Zhang Y, et al. Lignocellulose dissociation with biological pretreatment towards the biochemical platform: a review[J]. Mater Today Bio, 2022, 16: 100445. |
| [15] |
Sai Bharadwaj AVSL, Dev S, Zhuang JS, et al. Review of chemical pretreatment of lignocellulosic biomass using low-liquid and low-chemical catalysts for effective bioconversion[J]. Bioresour Technol, 2023, 368: 128339.
doi: 10.1016/j.biortech.2022.128339 URL |
| [16] |
Shan WW, Yan YL, Li YD, et al. Microbial tolerance engineering for boosting lactic acid production from lignocellulose[J]. Biotechnol Biofuels Bioprod, 2023, 16(1): 78.
doi: 10.1186/s13068-023-02334-y |
| [17] |
Balasundaram G, Banu R, Varjani S, et al. Recalcitrant compounds formation, their toxicity, and mitigation: key issues in biomass pretreatment and anaerobic digestion[J]. Chemosphere, 2022, 291(Pt 3): 132930.
doi: 10.1016/j.chemosphere.2021.132930 URL |
| [18] |
Guo HL, Zhao Y, Chang JS, et al. Inhibitor formation and detoxification during lignocellulose biorefinery: a review[J]. Bioresour Technol, 2022, 361: 127666.
doi: 10.1016/j.biortech.2022.127666 URL |
| [19] |
Ujor VC, Okonkwo CC. Microbial detoxification of lignocellulosic biomass hydrolysates: biochemical and molecular aspects, challenges, exploits and future perspectives[J]. Front Bioeng Biotechnol, 2022, 10: 1061667.
doi: 10.3389/fbioe.2022.1061667 URL |
| [20] |
Gutiérrez T, Ingram LO, Preston JF. Purification and characterization of a furfural reductase(FFR)from Escherichia coli strain LYO1- an enzyme important in the detoxification of furfural during ethanol production[J]. J Biotechnol, 2006, 121(2): 154-164.
pmid: 16111779 |
| [21] |
Miller EN, Jarboe LR, Yomano LP, et al. Silencing of NADPH-dependent oxidoreductase genes(yqhD and dkgA)in furfural-resistant ethanologenic Escherichia coli[J]. Appl Environ Microbiol, 2009, 75(13): 4315-4323.
doi: 10.1128/AEM.00567-09 URL |
| [22] |
Miller EN, Jarboe LR, Turner PC, et al. Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180[J]. Appl Environ Microbiol, 2009, 75(19): 6132-6141.
doi: 10.1128/AEM.01187-09 URL |
| [23] | Hadi SM, Shahabuddin, Rehman A. Specificity of the interaction of furfural with DNA[J]. Mutat Res, 1989, 225(3): 101-106. |
| [24] |
Khan QA, Shamsi FA, Hadi SM. Mutagenicity of furfural in plasmid DNA[J]. Cancer Lett, 1995, 89(1): 95-99.
pmid: 7882307 |
| [25] |
Wang JQ, Zhang Y, Chen YL, et al. Global regulator engineering significantly improved Escherichia coli tolerances toward inhibitors of lignocellulosic hydrolysates[J]. Biotechnol Bioeng, 2012, 109(12): 3133-3142.
doi: 10.1002/bit.v109.12 URL |
| [26] |
Farr SB, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium[J]. Microbiol Rev, 1991, 55(4): 561-585.
doi: 10.1128/mr.55.4.561-585.1991 pmid: 1779927 |
| [27] |
Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium[J]. Nat Rev Microbiol, 2013, 11(7): 443-454.
doi: 10.1038/nrmicro3032 pmid: 23712352 |
| [28] |
Boopathy R, Bokang H, Daniels L. Biotransformation of furfural and 5-hydroxymethyl furfural by enteric bacteria[J]. J Ind Microbiol, 1993, 11(3): 147-150.
doi: 10.1007/BF01583715 URL |
| [29] |
Miller EN, Turner PC, Jarboe LR, et al. Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol-producing Escherichia coli LY180[J]. Biotechnol Lett, 2010, 32(5): 661-667.
doi: 10.1007/s10529-010-0209-9 pmid: 20131081 |
| [30] |
Shahabuddin, Rahman A, Hadi SM. Specificity of the in vitro interaction of methylfurfural with DNA[J]. Mutagenesis, 1990, 5(2): 131-136.
pmid: 2140425 |
| [31] |
Jilani SB, Dev C, Eqbal D, et al. Deletion of pgi gene in E. coli increases tolerance to furfural and 5-hydroxymethyl furfural in media containing glucose-xylose mixture[J]. Microb Cell Fact, 2020, 19(1): 153.
doi: 10.1186/s12934-020-01414-0 pmid: 32723338 |
| [32] |
Turner PC, Miller EN, Jarboe LR, et al. YqhC regulates transcription of the adjacent Escherichia coli genes yqhD and dkgA that are involved in furfural tolerance[J]. J Ind Microbiol Biotechnol, 2011, 38(3): 431-439.
doi: 10.1007/s10295-010-0787-5 URL |
| [33] |
Wang X, Miller EN, Yomano LP, et al. Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate[J]. Appl Environ Microbiol, 2011, 77(15): 5132-5140.
doi: 10.1128/AEM.05008-11 URL |
| [34] |
Jilani SB, Prasad R, Yazdani SS. Overexpression of oxidoreductase YghA confers tolerance of furfural in ethanologenic Escherichia coli strain SSK42[J]. Appl Environ Microbiol, 2021, 87(23): e0185521.
doi: 10.1128/AEM.01855-21 URL |
| [35] |
Song HS, Jeon JM, Kim HJ, et al. Increase in furfural tolerance by combinatorial overexpression of NAD salvage pathway enzymes in engineered isobutanol-producing E. coli[J]. Bioresour Technol, 2017, 245(Pt B): 1430-1435.
doi: 10.1016/j.biortech.2017.05.197 URL |
| [36] |
Willson BJ, Herman R, Langer S, et al. Improved furfural tolerance in Escherichia coli mediated by heterologous NADH-dependent benzyl alcohol dehydrogenases[J]. Biochem J, 2022, 479(10): 1045-1058.
doi: 10.1042/BCJ20210811 URL |
| [37] |
Zheng HB, Wang X, Yomano LP, et al. Increase in furfural tolerance in ethanologenic Escherichia coli LY180 by plasmid-based expression of thyA[J]. Appl Environ Microbiol, 2012, 78(12): 4346-4352.
doi: 10.1128/AEM.00356-12 URL |
| [38] |
Geddes RD, Wang X, Yomano LP, et al. Polyamine transporters and polyamines increase furfural tolerance during xylose fermentation with ethanologenic Escherichia coli strain LY180[J]. Appl Environ Microbiol, 2014, 80(19): 5955-5964.
doi: 10.1128/AEM.01913-14 URL |
| [39] |
Glebes TY, Sandoval NR, Reeder PJ, et al. Genome-wide mapping of furfural tolerance genes in Escherichia coli[J]. PLoS One, 2014, 9(1): e87540.
doi: 10.1371/journal.pone.0087540 URL |
| [40] |
Tan ZG, Khakbaz P, Chen YX, et al. Engineering Escherichia coli membrane phospholipid head distribution improves tolerance and production of biorenewables[J]. Metab Eng, 2017, 44: 1-12.
doi: 10.1016/j.ymben.2017.08.006 URL |
| [41] | Kurgan G, Panyon LA, Rodriguez-Sanchez Y, et al. Bioprospecting of native efflux pumps to enhance furfural tolerance in ethanologenic Escherichia coli[J]. Appl Environ Microbiol, 2019, 85(6): e02985-e02918. |
| [42] |
Geiger LE, Morris DR. Polyamine deficiency reduces the rate of DNA replication fork movement in Escherichia coli[J]. Nature, 1978, 272(5655): 730-732.
doi: 10.1038/272730a0 |
| [43] |
Mills TY, Sandoval NR, Gill RT. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli[J]. Biotechnol Biofuels, 2009, 2: 26.
doi: 10.1186/1754-6834-2-26 |
| [44] |
Sá-Pessoa J, Paiva S, Ribas D, et al. SATP(YaaH), a succinate-acetate transporter protein in Escherichia coli[J]. Biochem J, 2013, 454(3): 585-595.
doi: 10.1042/BJ20130412 pmid: 23844911 |
| [45] |
Gimenez R, Nuñez MF, Badia J, et al. The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli[J]. J Bacteriol, 2003, 185(21): 6448-6455.
doi: 10.1128/JB.185.21.6448-6455.2003 pmid: 14563880 |
| [46] |
Roe AJ, McLaggan D, Davidson I, et al. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids[J]. J Bacteriol, 1998, 180(4): 767-772.
doi: 10.1128/JB.180.4.767-772.1998 pmid: 9473028 |
| [47] |
Walter A, Gutknecht J. Monocarboxylic acid permeation through lipid bilayer membranes[J]. J Membr Biol, 1984, 77(3): 255-264.
doi: 10.1007/BF01870573 URL |
| [48] |
Guan NZ, Liu L. Microbial response to acid stress: mechanisms and applications[J]. Appl Microbiol Biotechnol, 2020, 104(1): 51-65.
doi: 10.1007/s00253-019-10226-1 pmid: 31773206 |
| [49] | Sun YR. F1F0-ATPase functions under markedly acidic conditions in bacteria[M]// Regulation of Ca2+-ATPases, V-ATPases and F-ATPases. Cham: Springer, 2016: 459-468. |
| [50] |
Maurer LM, Yohannes E, Bondurant SS, et al. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12[J]. J Bacteriol, 2005, 187(1): 304-319.
doi: 10.1128/JB.187.1.304-319.2005 pmid: 15601715 |
| [51] |
Choi SH, Baumler DJ, Kaspar CW. Contribution of dps to acid stress tolerance and oxidative stress tolerance in Escherichia coli O157: H7[J]. Appl Environ Microbiol, 2000, 66(9): 3911-3916.
doi: 10.1128/AEM.66.9.3911-3916.2000 URL |
| [52] |
Cherrington CA, Hinton M, Chopra I. Effect of short-chain organic acids on macromolecular synthesis in Escherichia coli[J]. J Appl Bacteriol, 1990, 68(1): 69-74.
doi: 10.1111/jam.1990.68.issue-1 URL |
| [53] |
Roe AJ, O'Byrne C, McLaggan D, et al. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity[J]. Microbiology, 2002, 148(Pt 7): 2215-2222.
doi: 10.1099/00221287-148-7-2215 URL |
| [54] |
Brown JL, Ross T, McMeekin TA, et al. Acid habituation of Escherichia coli and the potential role of cyclopropane fatty acids in low pH tolerance[J]. Int J Food Microbiol, 1997, 37(2/3): 163-173.
doi: 10.1016/S0168-1605(97)00068-8 URL |
| [55] |
Shabala L, Ross T. Cyclopropane fatty acids improve Escherichia coli survival in acidified minimal media by reducing membrane permeability to H+ and enhanced ability to extrude H+[J]. Res Microbiol, 2008, 159(6): 458-461.
doi: 10.1016/j.resmic.2008.04.011 pmid: 18562182 |
| [56] | 李书廷, 洪坤强, 汪保卫, 等. 大肠杆菌乙酸耐受性菌株的构建及其耐受机制研究进展[J]. 微生物学通报, 2020, 47(12): 4250-4259. |
| Li ST, Hong KQ, Wang BW, et al. Advances in construction of acetic acid tolerance Escherichia coli[J]. Microbiol China, 2020, 47(12): 4250-4259. | |
| [57] | 郝雪雁, 刘梦晓, 韩紫依, 等. 大肠杆菌的耐酸机制及其改造研究进展[J]. 微生物学通报, 2023, 50(10):4667-4680. |
| Hao XY, Liu MX, Han ZY, et al. Advances in acid-resistant mechanisms and modifications of Escherichia coli[J]. Microbiol China, 2023, 50(10):4667-4680. | |
| [58] |
Mallick S, Das S. Acid-tolerant bacteria and prospects in industrial and environmental applications[J]. Appl Microbiol Biotechnol, 2023, 107(11): 3355-3374.
doi: 10.1007/s00253-023-12529-w |
| [59] |
Yang JH, Zhang J, Zhu ZM, et al. The challenges and prospects of Escherichia coli as an organic acid production host under acid stress[J]. Appl Microbiol Biotechnol, 2021, 105(21/22): 8091-8107.
doi: 10.1007/s00253-021-11577-4 |
| [60] |
Xu Y, Zhao Z, Tong WH, et al. An acid-tolerance response system protecting exponentially growing Escherichia coli[J]. Nat Commun, 2020, 11(1): 1496.
doi: 10.1038/s41467-020-15350-5 |
| [61] |
Kirkpatrick C, Maurer LM, Oyelakin NE, et al. Acetate and formate stress: opposite responses in the proteome of Escherichia coli[J]. J Bacteriol, 2001, 183(21): 6466-6477.
pmid: 11591692 |
| [62] | Kammel M, Pinske C, Sawers RG. FocA and its central role in fine-tuning pH homeostasis of enterobacterial formate metabolism[J]. Microbiology, 2022, 168(10). DOI: 10.1099/mic.0.001253. |
| [63] | Lin ZL, Li JH, Yan XF, et al. Engineering of the small noncoding RNA(sRNA)DsrA together with the sRNA chaperone Hfq enhances the acid tolerance of Escherichia coli[J]. Appl Environ Microbiol, 2021, 87(10): e02923-e02920. |
| [64] |
Hu WB, Tong YJ, Liu JJ, et al. Improving acid resistance of Escherichia coli base on the CfaS-mediated membrane engineering strategy derived from extreme acidophile[J]. Front Bioeng Biotechnol, 2023, 11: 1158931.
doi: 10.3389/fbioe.2023.1158931 URL |
| [65] |
Jiang ZM, Lu J, Tong YJ, et al. Enhancement of acid tolerance of Escherichia coli by introduction of molecule chaperone CbpA from extremophile[J]. World J Microbiol Biotechnol, 2023, 39(6): 158.
doi: 10.1007/s11274-023-03613-4 |
| [66] |
Yao XR, Liu P, Chen B, et al. Synthetic acid stress-tolerance modules improve growth robustness and lysine productivity of industrial Escherichia coli in fermentation at low pH[J]. Microb Cell Fact, 2022, 21(1): 68.
doi: 10.1186/s12934-022-01795-4 |
| [67] |
Zaldivar J, Martinez A, Ingram LO. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli[J]. Biotechnol Bioeng, 1999, 65(1): 24-33.
doi: 10.1002/(sici)1097-0290(19991005)65:1<24::aid-bit4>3.0.co;2-2 pmid: 10440668 |
| [68] |
Fitzgerald DJ, Stratford M, Gasson MJ, et al. Mode of antimicrobial action of vanillin against Escherichia coli, Lactobacillus plantarum and Listeria innocua[J]. J Appl Microbiol, 2004, 97(1): 104-113.
doi: 10.1111/j.1365-2672.2004.02275.x pmid: 15186447 |
| [69] | Pattrick CA, Webb JP, Green J, et al. Proteomic profiling, transcription factor modeling, and genomics of evolved tolerant strains elucidate mechanisms of vanillin toxicity in Escherichia coli[J]. mSystems, 2019, 4(4): e00163-e00119. |
| [70] |
Yu QH, Li YC, Wu B, et al. Novel mutagenesis and screening technologies for food microorganisms: advances and prospects[J]. Appl Microbiol Biotechnol, 2020, 104(4): 1517-1531.
doi: 10.1007/s00253-019-10341-z pmid: 31919586 |
| [71] |
Wu SR, Tian PF, Tan TW. Genomic landscapes of bacterial transposons and their applications in strain improvement[J]. Appl Microbiol Biotechnol, 2022, 106(19/20): 6383-6396.
doi: 10.1007/s00253-022-12170-z |
| [72] |
Zhang X, Zhang XF, Li HP, et al. Atmospheric and room temperature plasma(ARTP)as a new powerful mutagenesis tool[J]. Appl Microbiol Biotechnol, 2014, 98(12): 5387-5396.
doi: 10.1007/s00253-014-5755-y pmid: 24769904 |
| [73] |
Chen L, Xin QH, Ma LM, et al. Applications and research advance of genome shuffling for industrial microbial strains improvement[J]. World J Microbiol Biotechnol, 2020, 36(10): 158.
doi: 10.1007/s11274-020-02936-w |
| [74] | Yoon SH, Lee EG, Das A, et al. Enhanced vanillin production from recombinant E. coli using NTG mutagenesis and adsorbent resin[J]. Biotechnol Prog, 2007, 23(5): 1143-1148. |
| [75] |
Gao XX, Yang XF, Li JH, et al. Engineered global regulator H-NS improves the acid tolerance of E. coli[J]. Microb Cell Fact, 2018, 17(1): 118.
doi: 10.1186/s12934-018-0966-z |
| [76] |
Wang GL, Li Q, Zhang Z, et al. Recent progress in adaptive laboratory evolution of industrial microorganisms[J]. J Ind Microbiol Biotechnol, 2023, 50(1): kuac023.
doi: 10.1093/jimb/kuac023 URL |
| [77] |
Sandberg TE, Salazar MJ, Weng LL, et al. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology[J]. Metab Eng, 2019, 56: 1-16.
doi: S1096-7176(19)30153-3 pmid: 31401242 |
| [78] |
Lu Q, Zhou XL, Liu JZ. Adaptive laboratory evolution and shuffling of Escherichia coli to enhance its tolerance and production of astaxanthin[J]. Biotechnol Biofuels Bioprod, 2022, 15(1): 17.
doi: 10.1186/s13068-022-02118-w |
| [79] |
Seong W, Han GH, Lim HS, et al. Adaptive laboratory evolution of Escherichia coli lacking cellular byproduct formation for enhanced acetate utilization through compensatory ATP consumption[J]. Metab Eng, 2020, 62: 249-259.
doi: 10.1016/j.ymben.2020.09.005 URL |
| [80] |
Warner JR, Reeder PJ, Karimpour-Fard A, et al. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides[J]. Nat Biotechnol, 2010, 28(8): 856-862.
doi: 10.1038/nbt.1653 pmid: 20639866 |
| [81] |
Glebes TY, Sandoval NR, Gillis JH, et al. Comparison of genome-wide selection strategies to identify furfural tolerance genes in Escherichia coli[J]. Biotechnol Bioeng, 2015, 112(1): 129-140.
doi: 10.1002/bit.v112.1 URL |
| [82] | Chang DD, Islam ZU, Zheng JF, et al. Inhibitor tolerance and bioethanol fermentability of levoglucosan-utilizing Escherichia coli were enhanced by overexpression of stress-responsive gene ycfR: the proteomics-guided metabolic engineering[J]. Synth Syst Biotechnol, 2021, 6(4): 384-395. |
| [83] |
Forsberg KJ, Patel S, Witt E, et al. Identification of genes conferring tolerance to lignocellulose-derived inhibitors by functional selections in soil metagenomes[J]. Appl Environ Microbiol, 2015, 82(2): 528-537.
doi: 10.1128/AEM.02838-15 URL |
| [84] |
Gurdo N, Volke DC, McCloskey D, et al. Automating the design-build-test-learn cycle towards next-generation bacterial cell factories[J]. N Biotechnol, 2023, 74: 1-15.
doi: 10.1016/j.nbt.2023.01.002 URL |
| [85] |
Phaneuf PV, Zielinski DC, Yurkovich JT, et al. Escherichia coli data-driven strain design using aggregated adaptive laboratory evolution mutational data[J]. ACS Synth Biol, 2021, 10(12): 3379-3395.
doi: 10.1021/acssynbio.1c00337 URL |
| [86] |
Choi TR, Song HS, Han YH, et al. Enhanced tolerance to inhibitors of Escherichia coli by heterologous expression of cyclopropane-fatty acid-acyl-phospholipid synthase(cfa)from Halomonas socia[J]. Bioprocess Biosyst Eng, 2020, 43(5): 909-918.
doi: 10.1007/s00449-020-02287-8 |
| [87] | Choi KR, Shin JH, Cho JS, et al. Systems metabolic engineering of Escherichia coli[J]. EcoSal Plus, 2016, 7(1). DOI: 10.1128/ecosalplus.ESP-0010-2015. |
| [88] |
Choi KR, Jang WD, Yang D, et al. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering[J]. Trends Biotechnol, 2019, 37(8): 817-837.
doi: S0167-7799(19)30003-4 pmid: 30737009 |
| [89] |
Sandoval NR, Mills TY, Zhang M, et al. Elucidating acetate tolerance in E. coli using a genome-wide approach[J]. Metab Eng, 2011, 13(2): 214-224.
doi: 10.1016/j.ymben.2010.12.001 URL |
| [1] | CHEN Cai-ping, REN Hao, LONG Teng-fei, HE Bing, LU Zhao-xiang, SUN Jian. Research Advances in the Treatment of Inflammation Bowel Disease Using Escherichia coli Nissle 1917 [J]. Biotechnology Bulletin, 2023, 39(6): 109-118. |
| [2] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| [3] | WU Li-dan, RAN Xue-qin, NIU Xi, HUANG Shi-hui, LI Sheng, WANG Jia-fu. Genome Comparison and Virulence Factor Analysis of Pathogenic Escherichia coli from Porcine [J]. Biotechnology Bulletin, 2023, 39(12): 287-299. |
| [4] | LI Yi-ya, WU Yi-fan, DING Neng-shui, FAN Xiao-ping, CHEN Fan. Establishment of a Luciferase-assisted Quantitative Method for Measuring Ultrasonic Disruption of Escherichia coli Cells [J]. Biotechnology Bulletin, 2023, 39(12): 90-98. |
| [5] | LI Xin-yue, ZHOU Ming-hai, FAN Ya-chao, LIAO Sha, ZHANG Feng-li, LIU Chen-guang, SUN Yue, ZHANG Lin, ZHAO Xin-qing. Research Progress in the Improvement of Microbial Strain Tolerance and Efficiency of Biological Manufacturing Based on Transporter Engineering [J]. Biotechnology Bulletin, 2023, 39(11): 123-136. |
| [6] | SUN Yan-qiu, XIE Cai-yun, TANG Yue-qin. Construction and Mechanism Analysis of High-temperature Resistant Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(11): 226-237. |
| [7] | WANG Wen-tao, FENG Qi, LIU Chen-guang, BAI Feng-wu, ZHAO Xin-qing. Redox-sensitive Genetic Parts Improve the Tolerance of Yeast to Lignocellulosic Hydrolysate Inhibitors [J]. Biotechnology Bulletin, 2023, 39(11): 360-372. |
| [8] | LI Hai-li, LANG Li-min, ZHANG Qing-xian, YOU Yi, ZHU Wen-hao, WANG Zhi-fang, ZHANG Li-xian, WANG Ke-ling. Identification and Drug Resistance of Escherichia coli Simultaneously Producing Carbapenemase NDM-1 and NDM-5 [J]. Biotechnology Bulletin, 2022, 38(9): 106-115. |
| [9] | CHENG Shen-wei, ZHANG Ke-qiang, LIANG Jun-feng, LIU Fu-yuan, GAO Xing-liang, DU Lian-zhu. Establishment of a Triple Droplet Digital PCR Quantitative Detection Method for Typical Pathogenic Bacteria in Livestock and Poultry Manure [J]. Biotechnology Bulletin, 2022, 38(9): 271-280. |
| [10] | ZHAO Yan-kun, LIU Hui-min, MENG Lu, WANG Cheng, WANG Jia-qi, ZHENG Nan. Research Progress in Heteroresistance of Escherichia coli [J]. Biotechnology Bulletin, 2022, 38(9): 59-71. |
| [11] | GAO Wei-xin, HUANG Huo-qing, ZHAO Jing, ZHANG Xin, YANG Ning, YANG Hao-meng. Construction and Activity Verification of Ribonucleoprotein Complex for Gene Editing [J]. Biotechnology Bulletin, 2022, 38(8): 60-68. |
| [12] | CHEN Hong-yan, LI Xiao-er, LI Zhong-guang. Sugar Signaling and Its Role in Plant Response to Environmental Stress [J]. Biotechnology Bulletin, 2022, 38(7): 80-89. |
| [13] | SUN Man-luan, GE Sai, BU Jia, ZHU Zhuang-yan. Regulation Mechanism of Ribonucleases in Escherichia coli [J]. Biotechnology Bulletin, 2022, 38(3): 234-245. |
| [14] | WANG Kai-kai, WANG Xiao-lu, SU Xiao-yun, ZHANG Jie. Optimization and Application of Double-plasmid CRISPR-Cas9 System in Escherichia coli [J]. Biotechnology Bulletin, 2021, 37(12): 252-264. |
| [15] | ZHANG Dan, WANG Nan, LI Chao, XIE Qi, TANG San-yuan. Sweet Sorghum—a High Efficient and Quality Forage Crop [J]. Biotechnology Bulletin, 2019, 35(5): 2-8. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||