








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (11): 328-339.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0049
Previous Articles Next Articles
YAN Meng-yu( ), WEI Xiao-wei, CAO Jing, LAN Hai-yan(
), WEI Xiao-wei, CAO Jing, LAN Hai-yan( )
)
Received:2023-01-18
Online:2023-11-26
Published:2023-12-20
Contact:
LAN Hai-yan
E-mail:1610372465@qq.com;Lanhaiyan@xju.edu.cn
YAN Meng-yu, WEI Xiao-wei, CAO Jing, LAN Hai-yan. Cloning of Basic Helix-loop-helix(bHLH)Transcription Factor Gene SabHLH169 in Suaeda aralocaspica and Analysis of Its Resistances to Drought Stress[J]. Biotechnology Bulletin, 2023, 39(11): 328-339.
| 名称 Name | 上游引物序列 Forward primer(5'-3') | 下游引物序列 Reverse primer(5'-3') |
|---|---|---|
| SabHLH169(PCR) | ATGGCTACTCACTTGCAGCAGTTGCTTC | TCAGGCATTAGTTTTTGATTGAAGTAGTTGG |
| pSuper1300-SabHLH169 | GCGTCGACATGGCTACTCACTTGCAGCAGTTGCT | CGGGATCCTCAGGCATTAGTTTTTGATTGAAGTA |
| pMal-c2X-SabHLH169 | CGGGATCCATGGCTACTCACTTGCAGCAGTTGCTTC | GCGTCGACTCAGGCATTAGTTTTTGATTGAAGTAGTT |
| AtHPT | GGTCGCGGAGGCTATGGATGC | GCTTCTGCGGGCGATTTGTGT |
| AtRD22(qRT-PCR) | GACTTTCGATTTTACCGACGAG | CGCTACCGGTTTTACCTTTATG |
| AtDREB2A(qRT-PCR) | CGTTTCAGGATGAGATGTGTGA | CTCATCGTGCATATAAAACGCA |
| AtRD29A(qRT-PCR) | TTCTGTAAGGACGACGTTTACA | CGTACTCGTTACATCCTCTGTT |
| AtACTIN(qRT-PCR) | GGTAACATTGTGCTCAGTGGTGG | AACGACCTTAATCTTCATGCTGC |
| AtPEPC(qRT-PCR) | AGCCTTCAGGGAACCACAAT | CTCCAAAGACGGGTCGCATG |
| AtRubisco(qRT-PCR) | TGGCTTCCTCTATGCTCTCTTC | ACACTTGAGCGGAGTCGGTGCA |
| AtLhcb2(qRT-PCR) | ATGTTGGGTGCTCTCGGATG | CGCGTGGATCAAGTTAGGGT |
| AtRCA(qRT-PCR) | GGCCGCCGCAGTTTCCACCG | AAAGTTGTAGACACAGGTTCCA |
Table 1 Primer sequences used in this study
| 名称 Name | 上游引物序列 Forward primer(5'-3') | 下游引物序列 Reverse primer(5'-3') |
|---|---|---|
| SabHLH169(PCR) | ATGGCTACTCACTTGCAGCAGTTGCTTC | TCAGGCATTAGTTTTTGATTGAAGTAGTTGG |
| pSuper1300-SabHLH169 | GCGTCGACATGGCTACTCACTTGCAGCAGTTGCT | CGGGATCCTCAGGCATTAGTTTTTGATTGAAGTA |
| pMal-c2X-SabHLH169 | CGGGATCCATGGCTACTCACTTGCAGCAGTTGCTTC | GCGTCGACTCAGGCATTAGTTTTTGATTGAAGTAGTT |
| AtHPT | GGTCGCGGAGGCTATGGATGC | GCTTCTGCGGGCGATTTGTGT |
| AtRD22(qRT-PCR) | GACTTTCGATTTTACCGACGAG | CGCTACCGGTTTTACCTTTATG |
| AtDREB2A(qRT-PCR) | CGTTTCAGGATGAGATGTGTGA | CTCATCGTGCATATAAAACGCA |
| AtRD29A(qRT-PCR) | TTCTGTAAGGACGACGTTTACA | CGTACTCGTTACATCCTCTGTT |
| AtACTIN(qRT-PCR) | GGTAACATTGTGCTCAGTGGTGG | AACGACCTTAATCTTCATGCTGC |
| AtPEPC(qRT-PCR) | AGCCTTCAGGGAACCACAAT | CTCCAAAGACGGGTCGCATG |
| AtRubisco(qRT-PCR) | TGGCTTCCTCTATGCTCTCTTC | ACACTTGAGCGGAGTCGGTGCA |
| AtLhcb2(qRT-PCR) | ATGTTGGGTGCTCTCGGATG | CGCGTGGATCAAGTTAGGGT |
| AtRCA(qRT-PCR) | GGCCGCCGCAGTTTCCACCG | AAAGTTGTAGACACAGGTTCCA |

Fig. 1 Extraction of total RNA and PCR amplification of SabHLH169 ORF in S. aralocaspica A: Total RNA of S. aralocaspica seedlings. 1,2: Two samples of total RNA; B: PCR amplification of SabHLH169 ORF; M1, M2: DL2000, DL5000 DNA markers

Fig. 2 Analysis of amino acid sequences of SabHLH169 in S. aralocaspica N-terminal underline indicates bHLH-MYC_N domain; C-terminal underline indicates HLH domain

Fig. 3 Analysis of the phylogenetic tree and bHLH domain of SabHLH169 in S. aralocaspica A: Analysis of the phylogenetic tree of SabHLH169, which was constructed based on the NJ method using MEGA X software with 1 000 bootstraps. B: Multiple alignment of the bHLH motif of amino acid sequences between SabHLH169 and the closely related species. Different colored letters indicate different degrees of conservation(dark blue > pink red > light blue > grey; the dark background indicates sequence identical); the upper line indicates the HLH domain

Fig. 4 Analysis of the conserved motifs of SabHLH169 in C. quinoa and the closely related species A: Schematic diagram of the base enrichment of the top 10 motifs; boxes in different colors represent motif1-motif10; the height of the letter indicates the frequency of the amino acid, and the higher the height, the greater the frequency. B: Conserved motif analysis of bHLH transcription factors in different species
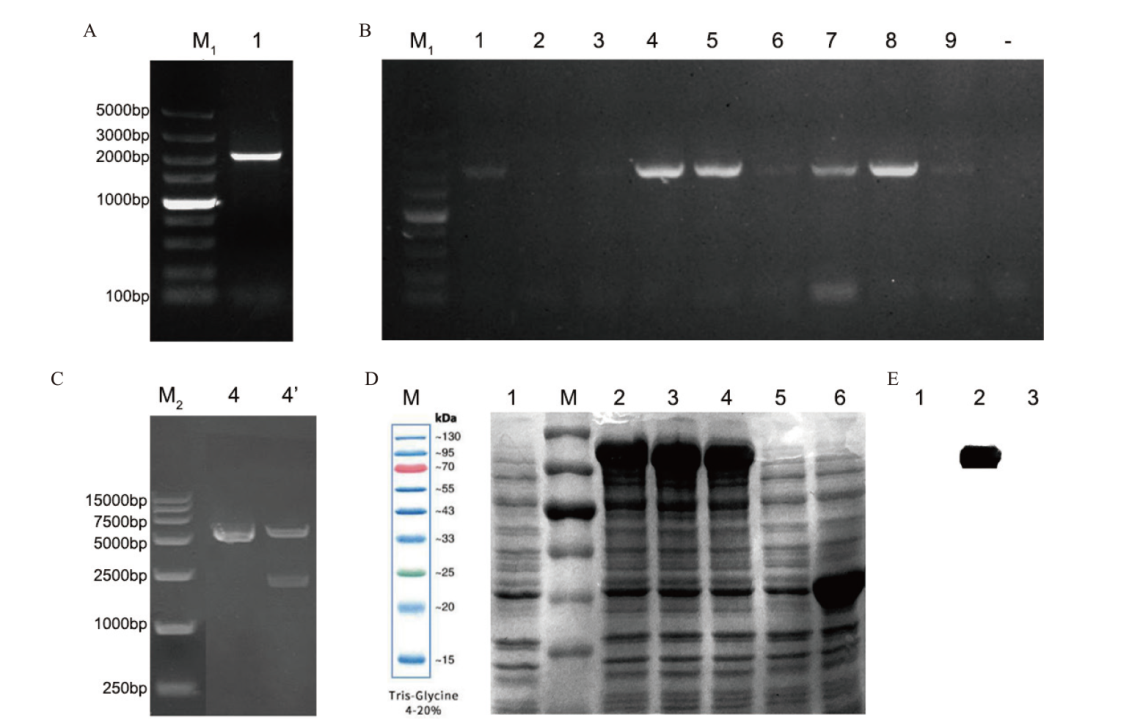
Fig. 5 Construction of procaryotic expression vector pMal-c2X -SabHLH169 and SDS-PAGE and Western blot analysis of SabHLH169 A: Amplification of SabHLH169. B: PCR identification of pMal-c2X-SabHLH169 transformed colonies; -: negative control. C: Double digestion of recombinant plasmid. D: SDS-PAGE results. M: Protein marker; 1: Before induction of pMal-c2X-SabHLH169; 2-4: after induction of pMal-c2X-SabHLH169(0.5 mmol/L, 0.8 mmol/L, 1 mmol/L IPTG); 5, 6: before and after induction of pMal-c2X. E: Western blot results; 1: After induction of pMal-c2X-SabHLH169(0.5 mmol/L IPTG); 2: before induction of pMal-c2X-SabHLH169; 3: before induction of pMal-c2X

Fig. 6 Construction of plant expression vector pSuper1300-SabHLH169 and identification of transgenic A. thaliana with SabHLH169 overexpression A: Amplification of SabHLH169. B: PCR identification of pSuper1300-SabHLH169 transformed colonies. C: Double digestion of recombinant plasmid. D: Identification of the T0 generation of transgenic A. thaliana overexpress SabHLH169. E, F: PCR and RT-qPCR identification of SabHLH169 in three T1 transgenic lines OE5, OE16, OE20. +: positive control; -: negative control. WT: Wild type A. thaliana; OE5, OE16 and OE20: three transgenic A. thaliana lines with SabHLH169 overexpression. M: DL 5000 DNA marker
| 株系 Strain | 阳性苗 Positive seedlings | 阴性苗 Negative seedlings | 卡方检验 Chi-square test(P-value) | 株系 Strain | 阳性苗 Positive seedling | 阴性苗 Negative seedling | 卡方检验 Chi-square test(P-value) | |
|---|---|---|---|---|---|---|---|---|
| OE1 | 134 | 37 | 0.310 | OE13 | 78 | 31 | 0.407 | |
| OE5 | 81 | 26 | 0.867 | OE14 | 64 | 27 | 0.304 | |
| OE6 | 70 | 33 | 0.099 | OE16 | 65 | 26 | 0.431 | |
| OE7 | 79 | 40 | 0.030 | OE17 | 62 | 26 | 0.325 | |
| OE8 | 78 | 32 | 0.322 | OE18 | 54 | 25 | 0.173 | |
| OE11 | 64 | 29 | 0.169 | OE19 | 54 | 26 | 0.121 | |
| OE12 | 61 | 24 | 0.491 | OE20 | 56 | 21 | 0.645 |
Table 2 Antibiotic screening of SabHLH169 transgenic A. thaliana T1 generation for 3∶1 segregation ratio lines
| 株系 Strain | 阳性苗 Positive seedlings | 阴性苗 Negative seedlings | 卡方检验 Chi-square test(P-value) | 株系 Strain | 阳性苗 Positive seedling | 阴性苗 Negative seedling | 卡方检验 Chi-square test(P-value) | |
|---|---|---|---|---|---|---|---|---|
| OE1 | 134 | 37 | 0.310 | OE13 | 78 | 31 | 0.407 | |
| OE5 | 81 | 26 | 0.867 | OE14 | 64 | 27 | 0.304 | |
| OE6 | 70 | 33 | 0.099 | OE16 | 65 | 26 | 0.431 | |
| OE7 | 79 | 40 | 0.030 | OE17 | 62 | 26 | 0.325 | |
| OE8 | 78 | 32 | 0.322 | OE18 | 54 | 25 | 0.173 | |
| OE11 | 64 | 29 | 0.169 | OE19 | 54 | 26 | 0.121 | |
| OE12 | 61 | 24 | 0.491 | OE20 | 56 | 21 | 0.645 |

Fig. 7 Seed germination of SabHLH169 transgenic A. thaliana under mannitol treatment A: Phenotype analysis. B: Statistics of seed germination percentage
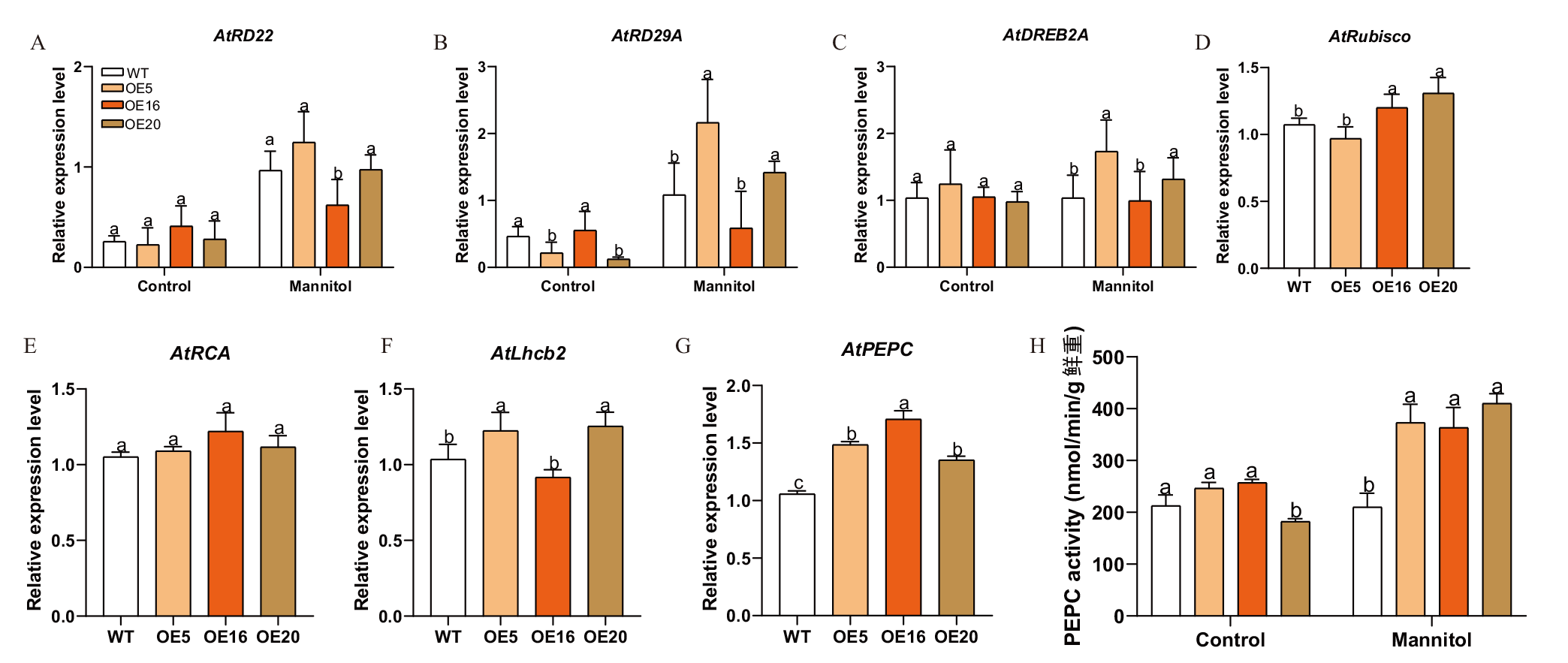
Fig. 8 Analysis of expression patterns of drought and photosynthesis related genes and PEPC enzyme activity in transgenic A. thaliana A-C: Expressions of drought stress-related genes in transgenic A.thaliana. D-G: Expressions of photosynthesis related genes. H: PEPC enzyme activity in transgenic A. thaliana. Different lowercase letters indicate significant differences in gene expression level at the same condition between different lines(P<0.05)
| [1] |
Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family[J]. Plant Cell, 2003, 15(8): 1749-1770.
doi: 10.1105/tpc.013839 pmid: 12897250 |
| [2] |
Li XX, Duan XP, Jiang HX, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis[J]. Plant Physiol, 2006, 141(4): 1167-1184.
doi: 10.1104/pp.106.080580 URL |
| [3] |
Sun X, Wang Y, Sui N. Transcriptional regulation of bHLH during plant response to stress[J]. Biochemical and Biophysical Research Communications, 2018, 503(2):397-401.
doi: S0006-291X(18)31625-5 pmid: 30057319 |
| [4] |
Castilhos G, Lazzarotto F, Spagnolo-Fonini L, et al. Possible roles of basic helix-loop-helix transcription factors in adaptation to drought[J]. Plant Sci, 2014, 223: 1-7.
doi: 10.1016/j.plantsci.2014.02.010 pmid: 24767109 |
| [5] |
Voznesenskaya EV, Franceschi VR, Kiirats O, et al. Kranz anatomy is not essential for terrestrial C4 plant photosynthesis[J]. Nature, 2001, 414(6863): 543-546.
doi: 10.1038/35107073 |
| [6] |
Edwards GE, Franceschi VR, Voznesenskaya EV. Single-cell C(4)photosynthesis versus the dual-cell(Kranz)paradigm[J]. Annu Rev Plant Biol, 2004, 55: 173-196.
pmid: 15377218 |
| [7] | Cao J, Cheng G, Wang L, et al. Genome-wide identification and analysis of the phosphoenolpyruvate carboxylase gene family in Suaeda aralocaspica, an annual halophyte with single-cellular C4 anatomy[J]. Frontiers in Plant Science, 2021, 12: 1603. |
| [8] | 曹婧. 非花环结构C4盐生植物异子蓬两种PEPC(PEPC-1和PEPC-2)基因的功能及其转录调控研究[D]. 乌鲁木齐: 新疆大学, 2019. |
| Cao J. Function and transcriptional regulation of two types of PEPC(PEPC-1 and PEPC-2)genes in Suaeda aralocaspica, a C4 halophyte with non-Kranz anatomy[D]. Urumqi: Xinjiang University, 2019. | |
| [9] |
Górska AM, Gouveia P, Borba AR, et al. ZmbHLH80 and ZmbHLH90 transcription factors act antagonistically and contribute to regulate PEPC1 cell-specific gene expression in maize[J]. The Plant Journal, 2019, 99(2): 270-285.
doi: 10.1111/tpj.14323 pmid: 30900785 |
| [10] |
Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR[J]. BioTechniques, 2005, 39(4): 519-525.
doi: 10.2144/000112010 pmid: 16235564 |
| [11] |
Zhang TT, Lv W, Zhang HS, et al. Genome-wide analysis of the basic Helix-Loop-Helix(bHLH)transcription factor family in maize[J]. BMC Plant Biol, 2018, 18(1): 235.
doi: 10.1186/s12870-018-1441-z |
| [12] |
Wang PF, Su L, Gao HH, et al. Genome-wide characterization of bHLH genes in grape and analysis of their potential relevance to abiotic stress tolerance and secondary metabolite biosynthesis[J]. Front Plant Sci, 2018, 9: 64.
doi: 10.3389/fpls.2018.00064 pmid: 29449854 |
| [13] |
Mao K, Dong Q, Li C, et al. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress[J]. Front Plant Sci, 2017, 8: 480.
doi: 10.3389/fpls.2017.00480 pmid: 28443104 |
| [14] |
Zhao PC, Li XX, Jia JT, et al. bHLH92 from sheepgrass acts as a negative regulator of anthocyanin/proanthocyandin accumulation and influences seed dormancy[J]. J Exp Bot, 2019, 70(1): 269-284.
doi: 10.1093/jxb/ery335 URL |
| [15] |
Wang L, Wang HL, Yin L, et al. Transcriptome assembly in Suaeda aralocaspica to reveal the distinct temporal gene/miRNA alterations between the dimorphic seeds during germination[J]. BMC Genomics, 2017, 18(1): 806.
doi: 10.1186/s12864-017-4209-1 pmid: 29052505 |
| [16] |
Ohashi-Ito K, Bergmann DC. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY[J]. Development, 2007, 134(16): 2959-2968.
pmid: 17626058 |
| [17] |
Wei SW, Xia R, Chen CX, et al. ZmbHLH124 identified in maize recombinant inbred lines contributes to drought tolerance in crops[J]. Plant Biotechnol, 2021, 19(10): 2069-2081.
doi: 10.1111/pbi.v19.10 URL |
| [18] |
Abe H, Yamaguchi-Shinozaki K, Urao T, et al. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression[J]. Plant Cell, 1997, 9(10): 1859-1868.
doi: 10.1105/tpc.9.10.1859 pmid: 9368419 |
| [19] |
Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1[J]. Genetics, 2000, 156(3): 1349-1362.
doi: 10.1093/genetics/156.3.1349 pmid: 11063707 |
| [20] |
Azpiroz-Leehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: going back and forth[J]. Trends Genet, 1997, 13(4): 152-156.
pmid: 9097726 |
| [21] |
Arnau J, Oliver RP. Inheritance and alteration of transforming DNA during an induced parasexual cycle in the imperfect fungus Cladosporium fulvum[J]. Curr Genet, 1993, 23(5): 508-511.
doi: 10.1007/BF00312643 URL |
| [22] |
Provvidenti R, Gonsalves D. Inheritance of resistance to cucumber mosaic virus in a transgenic tomato line expressing the coat protein gene of the white leaf strain[J]. J Hered, 1995, 86(2): 85-88.
doi: 10.1093/oxfordjournals.jhered.a111553 URL |
| [23] | 贾庆利. 外源基因在拟南芥转基因植株中的遗传研究[D]. 杨凌: 西北农林科技大学, 2004. |
| Jia QL. Genetic study of exogenous genes in transgenic Arabidopsis plants[D]. Yangling: Northwest A & F University, 2004. | |
| [24] |
Helm LT, Shi HY, Lerdau MT, et al. Solar-induced chlorophyll fluorescence and short-term photosynthetic response to drought[J]. Ecol Appl, 2020, 30(5): e02101.
doi: 10.1002/eap.v30.5 URL |
| [25] |
Sajad Majeed Z, Nancy G, Muslima N, et al. Impact of drought on photosynthesis: Molecular perspective[J]. Plant Gene, 2017, 11(2): 154-159.
doi: 10.1016/j.plgene.2017.04.003 URL |
| [26] |
Spreitzer RJ, Salvucci ME. Rubisco: structure, regulatory interactions, and possibilities for a better enzyme[J]. Annu Rev Plant Biol, 2002, 53: 449-475.
pmid: 12221984 |
| [27] | Tissue DT, Thomas RB, Strain BR. Long-term effects of elevated CO2 and nutrients on photosynthesis and rubisco in loblolly pine seedlings[J]. Plant Cell & Environment, 2010, 16(7): 859-865. |
| [28] |
Kurek I, Chang TK, Bertain SM, et al. Enhanced Thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress[J]. Plant Cell, 2007, 19(10): 3230-3241.
doi: 10.1105/tpc.107.054171 URL |
| [29] |
Demirevska-Kepova K, Holzer R, Simova-Stoilova L, et al. Heat stress effects on ribulose-1, 5-bisphosphate carboxylase/oxygenase, Rubisco binding protein and Rubisco activase in wheat leaves[J]. Biol Plant, 2005, 49(4): 521-525.
doi: 10.1007/s10535-005-0045-2 URL |
| [30] |
Yin ZT, Zhang ZL, Deng DX, et al. Characterization of Rubisco activase genes in maize: an α-isoform gene functions alongside a β-isoform gene[J]. Plant Physiol, 2014, 164(4): 2096-2106.
doi: 10.1104/pp.113.230854 pmid: 24510763 |
| [31] |
Sharkey TD, Badger MR, von Caemmerer S, et al. Increased heat sensitivity of photosynthesis in tobacco plants with reduced Rubisco activase[J]. Photosynth Res, 2001, 67(1): 147-156.
doi: 10.1023/A:1010633823747 URL |
| [32] |
Wang D, Li XF, Zhou ZJ, et al. Two Rubisco activase isoforms may play different roles in photosynthetic heat acclimation in the rice plant[J]. Physiol Plant, 2010, 139(1): 55-67.
doi: 10.1111/j.1399-3054.2009.01344.x pmid: 20059735 |
| [33] | Jackowski G, Kacprzak K, Jansson S. Identification of Lhcb1/Lhcb2/Lhcb3 heterotrimers of the main light-harvesting chlorophyll a/b-protein complex of Photosystem II(LHC II)[J]. Biochim Biophys Acta BBA Bioenerg, 2001, 1504(2/3): 340-345. |
| [34] |
O’Leary B, Park J, Plaxton WC. The remarkable diversity of plant PEPC(phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs[J]. Biochem J, 2011, 436(1): 15-34.
doi: 10.1042/BJ20110078 URL |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [3] | LIU Wen-jin, MA Rui, LIU Sheng-yan, YANG Jiang-wei, ZHANG Ning, SI Huai-jun. Cloning of StCIPK11 Gene and Analysis of Its Response to Drought Stress in Solanum tuberosum [J]. Biotechnology Bulletin, 2023, 39(9): 147-155. |
| [4] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [5] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [6] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [7] | DING Kai-xin, WANG Li-chun, TIAN Guo-kui, WANG Hai-yan, LI Feng-yun, PAN Yang, PANG Ze, SHAN Ying. Research Progress in Uniconazole Alleviating Plant Drought Damage [J]. Biotechnology Bulletin, 2023, 39(6): 1-11. |
| [8] | WANG Chun-yu, LI Zheng-jun, WANG Ping, ZHANG Li-xia. Physiological and Biochemical Analysis of Drought Resistance in Sorghum Cuticular Wax-deficient Mutant sb1 [J]. Biotechnology Bulletin, 2023, 39(5): 160-167. |
| [9] | WANG Hai-long, LI Yu-qian, WANG Bo, XING Guo-fang, ZHANG Jie-wei. Isolation and Expression Analysis of SiMAPK3 in Setaria italica L. [J]. Biotechnology Bulletin, 2023, 39(3): 123-132. |
| [10] | WANG Qi, HU Zhe, FU Wei, LI Guang-zhe, HAO Lin. Regulation of Burkholderia sp. GD17 on the Drought Tolerance of Cucumber Seedlings [J]. Biotechnology Bulletin, 2023, 39(3): 163-175. |
| [11] | LIU Cheng-xia, SUN Zong-yan, LUO Yun-bo, ZHU Hong-liang, QU Gui-qin. Multifaceted Roles of bHLH Phosphorylation in Regulation of Plant Physiological Functions [J]. Biotechnology Bulletin, 2023, 39(3): 26-34. |
| [12] | YU Bo, QIN Xiao-hui, ZHAO Yang. Mechanisms of Plant Sensing Drought Signals [J]. Biotechnology Bulletin, 2023, 39(11): 6-17. |
| [13] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| [14] | FENG Ce-ting, JIANG Lyu, LIU Xing-ying, LUO Le, PAN Hui-tang, ZHANG Qi-xiang, YU Chao. Identification of the NAC Gene Family in Rosa persica and Response Analysis Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 283-296. |
| [15] | AN Chang, LU Lin, SHEN Meng-qian, CHEN Sheng-zhen, YE Kang-zhuo, QIN Yuan, ZHENG Ping. Research Progress of bHLH Gene Family in Plants and Its Application Prospects in Medical Plants [J]. Biotechnology Bulletin, 2023, 39(10): 1-16. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||