








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (4): 24-37.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1248
Previous Articles Next Articles
Received:2022-10-10
Online:2023-04-26
Published:2023-05-16
YU Hui-li, LI Ai-tao. Application of Cytochrome P450 in the Biosynthesis of Flavors and Fragrances[J]. Biotechnology Bulletin, 2023, 39(4): 24-37.
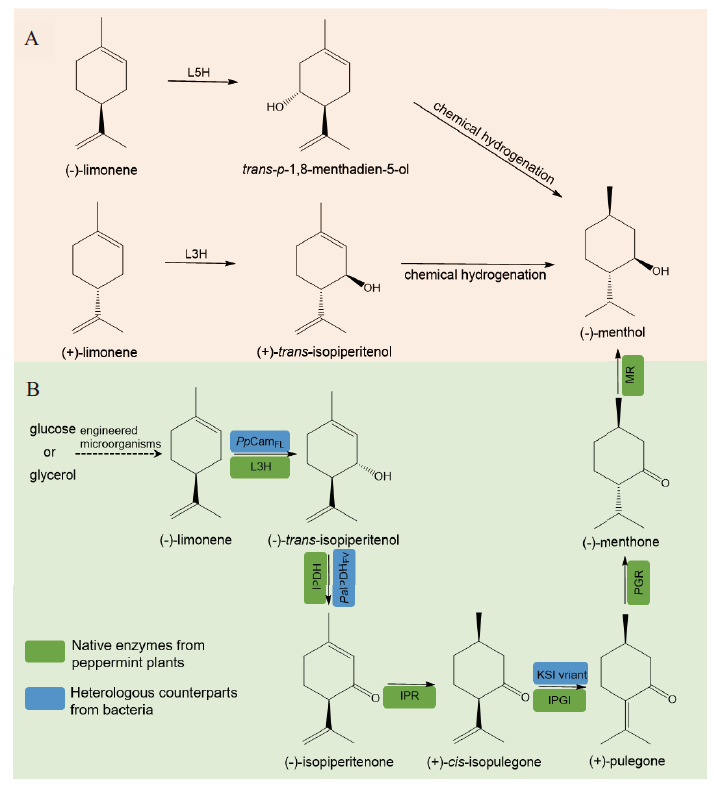
Fig. 5 P450 involved in the biotechnological-chemical processes(A)versus the de novo biosynthetic pathway for(-)-menthol production L5H: Limonene-5-hydroxylase; L3H: limonene-3-hydroxylase; IPDH:(-)-trans-isopiperitenol dehydrogenase; IPR:(-)-isopiperitenone reductase; IPGI:(+)-cis-isopulegone isomerase; PGR:(+)-pulegone reductase; MR:(-)-menthone reductase; PpCamFL: P450cam_Y96F/V247L variant from Pseudomonas putida; PaIPDHFV: IPDH_E95F/Y199V variant from Pseudomonas aeruginosa; KSI variant: Δ5-3-ketosteroid isomerase KSI_V88I/L99V/V101A/D103S variant from P. putida

Fig. 6 P450 involved in the de novo biosynthesis of S-(-)-perillyl alcohol AACT: Acetyl-CoA C-acetyltransferase; HMGS: hydroxymethylglutaryl-CoA synthase; HMGR: hydroxymethylglutaryl-CoA reductase; MVK: mevalonate kinase; PMVK: phosphomevalonate kinase; MVD: diphosphomevalonate decarboxylase; IDI: isopentenyl-diphosphate delta-isomerase; GPPS: GPP synthase; LS: limonene synthase

Fig. 7 P450 involved in the biotransformation of(+)-valencene(A)and de novo biosynthesis of(+)-nootkatone(B) FPPS: FPP synthase; CnVS: valencene synthase from Chamaecyparis nootkatensis; ADH: alcohol dehydrogenase
| [1] |
Nelson DR, Koymans L, Kamataki T, et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature[J]. Pharmacogenetics, 1996, 6(1): 1-42.
doi: 10.1097/00008571-199602000-00002 pmid: 8845856 |
| [2] |
Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes[J]. J Biol Chem, 1964, 239(7): 2370-2378.
doi: 10.1016/S0021-9258(20)82244-3 URL |
| [3] |
Nelson DR. Cytochrome P450 diversity in the tree of life[J]. Biochim Biophys Acta Proteins Proteom, 2018, 1866(1): 141-154.
doi: 10.1016/j.bbapap.2017.05.003 URL |
| [4] |
Miles CS, Ost TWB, Noble MA, et al. Protein engineering of cytochromes P-450[J]. Biochim Biophys Acta Protein Struct Mol Enzymol, 2000, 1543(2): 383-407.
doi: 10.1016/S0167-4838(00)00236-3 URL |
| [5] | Werck-Reichhart D, Feyereisen R. Cytochromes P450: a success story[J]. Genome Biol, 2000, 1(6): REVIEWS3003. |
| [6] |
Urlacher VB, Eiben S. Cytochrome P450 monooxygenases: perspectives for synthetic application[J]. Trends Biotechnol, 2006, 24(7): 324-330.
doi: 10.1016/j.tibtech.2006.05.002 pmid: 16759725 |
| [7] |
Graham SE, Peterson JA. How similar are P450s and what can their differences teach us?[J]. Arch Biochem Biophys, 1999, 369(1): 24-29.
pmid: 10462437 |
| [8] |
Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity[J]. Chem Res Toxicol, 2001, 14(6): 611-650.
doi: 10.1021/tx0002583 pmid: 11409933 |
| [9] |
Rittle J, Green MT. Cytochrome P450 compound I: capture, characterization, and C-H bond activation kinetics[J]. Science, 2010, 330(6006): 933-937.
doi: 10.1126/science.1193478 pmid: 21071661 |
| [10] |
Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome p450 enzymes[J]. Chem Rev, 2004, 104(9): 3947-3980.
doi: 10.1021/cr020443g pmid: 15352783 |
| [11] |
Li Z, Jiang YY, Guengerich FP, et al. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications[J]. J Biol Chem, 2020, 295(3): 833-849.
doi: 10.1074/jbc.REV119.008758 pmid: 31811088 |
| [12] |
Whitehouse CJC, Bell SG, Wong LL. P450(BM3)(CYP102A1): connecting the dots[J]. Chem Soc Rev, 2012, 41(3): 1218-1260.
doi: 10.1039/C1CS15192D URL |
| [13] |
Degtyarenko KN, Kulikova TA. Evolution of bioinorganic motifs in P450-containing systems[J]. Biochem Soc Trans, 2001, 29(Pt 2): 139-147.
doi: 10.1042/bst0290139 URL |
| [14] |
Hannemann F, Bichet A, Ewen KM, et al. Cytochrome P450 systems—biological variations of electron transport chains[J]. Biochim Biophys Acta, 2007, 1770(3): 330-344.
pmid: 16978787 |
| [15] |
Sakaki T. Practical application of cytochrome P450[J]. Biol Pharm Bull, 2012, 35(6): 844-849.
pmid: 22687473 |
| [16] | Xu LH, Du YL. Rational and semi-rational engineering of cytochrome P450s for biotechnological applications[J]. Synth Syst Biotechnol, 2018, 3(4): 283-290. |
| [17] |
Roberts GA, Çelik A, Hunter DJB, et al. A self-sufficient cytochrome P450 with a primary structural organization that includes a flavin domain and a[2Fe-2S]redox center[J]. J Biol Chem, 2003, 278(49): 48914-48920.
doi: 10.1074/jbc.M309630200 pmid: 14514666 |
| [18] |
Daiber A, Shoun H, Ullrich V. Nitric oxide reductase(P450nor)from Fusarium oxysporum[J]. J Inorg Biochem, 2005, 99(1): 185-193.
doi: 10.1016/j.jinorgbio.2004.09.018 URL |
| [19] |
Choi KY, Jung E, Jung DH, et al. Engineering of daidzein 3'-hydroxylase P450 enzyme into catalytically self-sufficient cytochrome P450[J]. Microb Cell Fact, 2012, 11: 81.
doi: 10.1186/1475-2859-11-81 |
| [20] |
Li SY, Podust LM, Sherman DH. Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain[J]. J Am Chem Soc, 2007, 129(43): 12940-12941.
doi: 10.1021/ja075842d pmid: 17915876 |
| [21] |
Li SY, Du L, Bernhardt R. Redox partners: function modulators of bacterial P450 enzymes[J]. Trends Microbiol, 2020, 28(6): 445-454.
doi: S0966-842X(20)30048-2 pmid: 32396826 |
| [22] |
Coon MJ. Cytochrome P450: nature's most versatile biological catalyst[J]. Annu Rev Pharmacol Toxicol, 2005, 45: 1-25.
pmid: 15832443 |
| [23] |
Guengerich FP, Munro AW. Unusual cytochrome p450 enzymes and reactions[J]. J Biol Chem, 2013, 288(24): 17065-17073.
doi: 10.1074/jbc.R113.462275 pmid: 23632016 |
| [24] |
Lamb DC, Waterman MR. Unusual properties of the cytochrome P450 superfamily[J]. Philos Trans R Soc Lond B Biol Sci, 2013, 368(1612): 20120434.
doi: 10.1098/rstb.2012.0434 URL |
| [25] |
Zhang XW, Li SY. Expansion of chemical space for natural products by uncommon P450 reactions[J]. Nat Prod Rep, 2017, 34(9): 1061-1089.
doi: 10.1039/c7np00028f pmid: 28770915 |
| [26] |
Podust LM, Sherman DH. Diversity of P450 enzymes in the biosynthesis of natural products[J]. Nat Prod Rep, 2012, 29(10): 1251-1266.
doi: 10.1039/c2np20020a pmid: 22820933 |
| [27] |
Rudolf JD, Chang CY, Ma M, et al. Cytochromes P450 for natural product biosynthesis in Streptomyces: sequence, structure, and function[J]. Nat Prod Rep, 2017, 34(9): 1141-1172.
doi: 10.1039/C7NP00034K URL |
| [28] |
Bernhardt R, Urlacher VB. Cytochromes P450 as promising catalysts for biotechnological application: chances and limitations[J]. Appl Microbiol Biotechnol, 2014, 98(14): 6185-6203.
doi: 10.1007/s00253-014-5767-7 pmid: 24848420 |
| [29] |
Yasuda K, Sugimoto H, Hayashi K, et al. Protein engineering of CYP105s for their industrial uses[J]. Biochim Biophys Acta Proteins Proteom, 2018, 1866(1): 23-31.
doi: 10.1016/j.bbapap.2017.05.014 URL |
| [30] |
Paddon CJ, Westfall PJ, Pitera DJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532.
doi: 10.1038/nature12051 |
| [31] |
McLean KJ, Hans M, Meijrink B, et al. Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum[J]. Proc Natl Acad Sci USA, 2015, 112(9): 2847-2852.
doi: 10.1073/pnas.1419028112 URL |
| [32] |
Urlacher V B, Girhard M. Cytochrome P450 monooxygenases: an update on perspectives for synthetic application[J]. Trends Biotechnol, 2012, 30(1): 26-36.
doi: 10.1016/j.tibtech.2011.06.012 pmid: 21782265 |
| [33] |
Girvan HM, Munro AW. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology[J]. Curr Opin Chem Biol, 2016, 31: 136-145.
doi: 10.1016/j.cbpa.2016.02.018 pmid: 27015292 |
| [34] |
Wei YF, Ang EL, Zhao HM. Recent developments in the application of P450 based biocatalysts[J]. Curr Opin Chem Biol, 2018, 43: 1-7.
doi: S1367-5931(17)30120-5 pmid: 29100098 |
| [35] |
Kolwek J, Behrens C, Linke D, et al. Cell-free one-pot conversion of(+)-valencene to(+)-nootkatone by a unique dye-decolorizing peroxidase combined with a laccase from Funalia trogii[J]. J Ind Microbiol Biotechnol, 2018, 45(2): 89-101.
doi: 10.1007/s10295-017-1998-9 URL |
| [36] | Surburg H, Panten J. Natural raw materials in the flavor and fragrance industry[M]// Common Fragrance and Flavor Materials. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA, 2016: 193-264. |
| [37] |
Rottava I, Cortina PF, Martello E, et al. Optimization of α-terpineol production by the biotransformation of R-(+)-limonene and(-)-β-pinene[J]. Appl Biochem Biotechnol, 2011, 164(4): 514-523.
doi: 10.1007/s12010-010-9153-3 pmid: 21234702 |
| [38] |
Croteau RB, Davis EM, Ringer KL, et al. (-)-Menthol biosynthesis and molecular genetics[J]. Naturwissenschaften, 2005, 92(12): 562-577.
doi: 10.1007/s00114-005-0055-0 URL |
| [39] |
Kamatou GPP, Vermaak I, Viljoen AM, et al. Menthol: a simple monoterpene with remarkable biological properties[J]. Phytochemistry, 2013, 96: 15-25.
doi: 10.1016/j.phytochem.2013.08.005 pmid: 24054028 |
| [40] | Schempp FM, Strobel I, Etschmann MMW, et al. Identification of fungal limonene-3-hydroxylase for biotechnological menthol production[J]. Appl Environ Microbiol, 2021, 87(10): e02873-e02820. |
| [41] |
Lupien S, Karp F, Wildung M, et al. Regiospecific cytochrome P450 limonene hydroxylases from mint(Mentha)species: cDNA isolation, characterization, and functional expression of(-)-4S-limonene-3-hydroxylase and(-)-4S-limonene-6-hydroxylase[J]. Arch Biochem Biophys, 1999, 368(1): 181-192.
doi: 10.1006/abbi.1999.1298 pmid: 10415126 |
| [42] |
Schalk M, Croteau R. A single amino acid substitution(F363I)converts the regiochemistry of the spearmint(-)-limonene hydroxylase from a C6- to a C3-hydroxylase[J]. Proc Natl Acad Sci USA, 2000, 97(22): 11948-11953.
doi: 10.1073/pnas.97.22.11948 pmid: 11050228 |
| [43] | Bell SG, Sowden RJ, Wong LL. Engineering the haem monooxygenase cytochrome P450cam for monoterpene oxidation[J]. Chem Commun, 2001(7): 635-636. |
| [44] | Shou C, Zheng YC, Zhan JR, et al. Removing the obstacle to(-)-menthol biosynthesis by building a microbial cell factory of(+)- cis-isopulegone from(-)-limonene[J]. ChemSusChem, 2022, 15(9): e202101741. |
| [45] |
Currin A, Dunstan MS, Johannissen LO, et al. Engineering the “missing link” in biosynthetic(-)-menthol production: bacterial isopulegone isomerase[J]. ACS Catal, 2018, 8(3): 2012-2020.
doi: 10.1021/acscatal.7b04115 URL |
| [46] |
Willrodt C, Hoschek A, Bühler B, et al. Coupling limonene formation and oxyfunctionalization by mixed-culture resting cell fermentation[J]. Biotechnol Bioeng, 2015, 112(9): 1738-1750.
doi: 10.1002/bit.25592 pmid: 25786991 |
| [47] |
Zhang X, Liu X, Meng Y, et al. Combinatorial engineering of Saccharomyces cerevisiae for improving limonene production[J]. Biochem Eng J, 2021, 176: 108155.
doi: 10.1016/j.bej.2021.108155 URL |
| [48] |
Dusséaux S, Wajn WT, Liu YX, et al. Transforming yeast peroxisomes into microfactories for the efficient production of high-value isoprenoids[J]. Proc Natl Acad Sci USA, 2020, 117(50): 31789-31799.
doi: 10.1073/pnas.2013968117 pmid: 33268495 |
| [49] |
Cao X, Lv YB, Chen J, et al. Metabolic engineering of oleaginous yeast Yarrowia lipolytica for limonene overproduction[J]. Biotechnol Biofuels, 2016, 9: 214.
doi: 10.1186/s13068-016-0626-7 URL |
| [50] |
Pina LTS, Serafini MR, Oliveira MA, et al. Carvone and its pharmacological activities: a systematic review[J]. Phytochemistry, 2022, 196: 113080.
doi: 10.1016/j.phytochem.2021.113080 URL |
| [51] |
Morrish JLE, Brennan ET, Dry HC, et al. Enhanced bioproduction of carvone in a two-liquid-phase partitioning bioreactor with a highly hydrophobic biocatalyst[J]. Biotechnol Bioeng, 2008, 101(4): 768-775.
doi: 10.1002/bit.21941 pmid: 18478563 |
| [52] |
Davis EM, Ringer KL, McConkey ME, et al. Monoterpene metabolism. Cloning, expression, and characterization of menthone reductases from peppermint[J]. Plant Physiol, 2005, 137(3): 873-881.
doi: 10.1104/pp.104.053306 pmid: 15728344 |
| [53] |
Yoshida E, Kojima M, Suzuki M, et al. Increased carvone production in Escherichia coli by balancing limonene conversion enzyme expression via targeted quantification concatamer proteome analysis[J]. Sci Rep, 2021, 11(1): 22126.
doi: 10.1038/s41598-021-01469-y pmid: 34764337 |
| [54] |
Zhang LL, Fan G, Li X, et al. Identification of functional genes associated with the biotransformation of limonene to trans-dihydrocarvone in Klebsiella sp. O852[J]. J Sci Food Agric, 2022, 102(8): 3297-3307.
doi: 10.1002/jsfa.v102.8 URL |
| [55] |
Sun C, Dong XJ, Zhang RB, et al. Effectiveness of recombinant Escherichia coli on the production of(R)-(+)-perillyl alcohol[J]. BMC Biotechnol, 2021, 21: 3.
doi: 10.1186/s12896-020-00662-7 |
| [56] |
Mau CJD, Karp F, Ito M, et al. A candidate cDNA clone for(-)-limonene-7-hydroxylase from Perilla frutescens[J]. Phytochemistry, 2010, 71(4): 373-379.
doi: 10.1016/j.phytochem.2009.12.002 URL |
| [57] |
Fujiwara Y, Ito M. Molecular cloning and characterization of a Perilla frutescens cytochrome P450 enzyme that catalyzes the later steps of perillaldehyde biosynthesis[J]. Phytochemistry, 2017, 134: 26-37.
doi: S0031-9422(16)30259-X pmid: 27890582 |
| [58] |
van Beilen JB, Holtackers R, Lüscher D, et al. Biocatalytic production of perillyl alcohol from limonene by using a novel Mycobacterium sp. cytochrome P450 alkane hydroxylase expressed in Pseudomonas putida[J]. Appl Environ Microbiol, 2005, 71(4): 1737-1744.
doi: 10.1128/AEM.71.4.1737-1744.2005 URL |
| [59] |
Cornelissen S, Liu SS, Deshmukh AT, et al. Cell physiology rather than enzyme kinetics can determine the efficiency of cytochrome P450-catalyzed C-H-oxyfunctionalization[J]. J Ind Microbiol Biotechnol, 2011, 38(9): 1359-1370.
doi: 10.1007/s10295-010-0919-y URL |
| [60] |
Cornelissen S, Julsing MK, Volmer J, et al. Whole-cell-based CYP153A6-catalyzed(S)-limonene hydroxylation efficiency depends on host background and profits from monoterpene uptake via AlkL[J]. Biotechnol Bioeng, 2013, 110(5): 1282-1292.
doi: 10.1002/bit.24801 pmid: 23239244 |
| [61] |
Seifert A, Antonovici M, Hauer B, et al. An efficient route to selective bio-oxidation catalysts: an iterative approach comprising modeling, diversification, and screening, based on CYP102A1[J]. Chembiochem, 2011, 12(9): 1346-1351.
doi: 10.1002/cbic.201100067 pmid: 21591046 |
| [62] |
Alonso-Gutierrez J, Chan R, Batth TS, et al. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production[J]. Metab Eng, 2013, 19: 33-41.
doi: 10.1016/j.ymben.2013.05.004 pmid: 23727191 |
| [63] |
Bicas JL, Barros FF, Wagner R, et al. Optimization of R-(+)-α-terpineol production by the biotransformation of R-(+)-limonene[J]. J Ind Microbiol Biotechnol, 2008, 35(9): 1061-1070.
doi: 10.1007/s10295-008-0383-0 URL |
| [64] |
Tai YN, Xu M, Ren JN, et al. Optimisation of α-terpineol production by limonene biotransformation using Penicillium digitatum DSM 62840[J]. J Sci Food Agric, 2016, 96(3): 954-961.
doi: 10.1002/jsfa.7171 URL |
| [65] |
Molina G, Pessôa MG, Bicas JL, et al. Optimization of limonene biotransformation for the production of bulk amounts of α-terpineol[J]. Bioresour Technol, 2019, 294: 122180.
doi: 10.1016/j.biortech.2019.122180 URL |
| [66] |
Zhang LL, Huang W, Zhang YY, et al. Genomic and transcriptomic study for screening genes involved in the limonene biotransformation of Penicillium digitatum DSM 62840[J]. Front Microbiol, 2020, 11: 744.
doi: 10.3389/fmicb.2020.00744 URL |
| [67] |
Fraatz MA, Berger RG, Zorn H. Nootkatone—a biotechnological challenge[J]. Appl Microbiol Biotechnol, 2009, 83(1): 35-41.
doi: 10.1007/s00253-009-1968-x pmid: 19333595 |
| [68] |
Zhu BC, Henderson G, Chen F, et al. Nootkatone is a repellent for Formosan subterranean termite(Coptotermes formosanus)[J]. J Chem Ecol, 2001, 27(3): 523-531.
pmid: 11441443 |
| [69] |
Clarkson TC, Janich AJ, Sanchez-Vargas I, et al. Nootkatone is an effective repellent against Aedes aegypti and Aedes albopictus[J]. Insects, 2021, 12(5): 386.
doi: 10.3390/insects12050386 URL |
| [70] |
Waltz E. A biotech insect repellent, safe enough to eat[J]. Nat Biotechnol, 2020, 38(12): 1368-1369.
doi: 10.1038/s41587-020-00760-z pmid: 33273737 |
| [71] |
Seifert A, Vomund S, Grohmann K, et al. Rational design of a minimal and highly enriched CYP102A1 mutant library with improved regio-, stereo- and chemoselectivity[J]. Chembiochem, 2009, 10(5): 853-861.
doi: 10.1002/cbic.200800799 pmid: 19222039 |
| [72] |
Girhard M, Machida K, Itoh M, et al. Regioselective biooxidation of(+)-valencene by recombinant E. coli expressing CYP109B1 from Bacillus subtilis in a two-liquid-phase system[J]. Microb Cell Fact, 2009, 8: 36.
doi: 10.1186/1475-2859-8-36 pmid: 19591681 |
| [73] | Kokorin A, Urlacher VB. Artificial fusions between P450 BM3 and an alcohol dehydrogenase for efficient(+)-nootkatone production[J]. ChemBioChem, 2022, 23(12): e202200065. |
| [74] |
Gavira C, Höfer R, Lesot A, et al. Challenges and pitfalls of P450-dependent(+)-valencene bioconversion by Saccharomyces cerevisiae[J]. Metab Eng, 2013, 18: 25-35.
doi: 10.1016/j.ymben.2013.02.003 URL |
| [75] |
Cankar K, van Houwelingen A, Bosch D, et al. A chicory cytochrome P450 mono-oxygenase CYP71AV8 for the oxidation of(+)-valencene[J]. FEBS Lett, 2011, 585(1): 178-182.
doi: 10.1016/j.febslet.2010.11.040 URL |
| [76] |
Cankar K, van Houwelingen A, Goedbloed M, et al. Valencene oxidase CYP706M1 from Alaska cedar(Callitropsis nootkatensis)[J]. FEBS Lett, 2014, 588(6): 1001-1007.
doi: 10.1016/j.febslet.2014.01.061 URL |
| [77] |
Guo X, Sun J, Li D, et al. Heterologous biosynthesis of(+)-nootkatone in unconventional yeast Yarrowia lipolytica[J]. Biochem Eng J, 2018, 137: 125-131.
doi: 10.1016/j.bej.2018.05.023 URL |
| [78] |
Takahashi S, Yeo YS, Zhao YX, et al. Functional characterization of premnaspirodiene oxygenase, a cytochrome P450 catalyzing regio- and stereo-specific hydroxylations of diverse sesquiterpene substrates[J]. J Biol Chem, 2007, 282(43): 31744-31754.
doi: 10.1074/jbc.M703378200 pmid: 17715131 |
| [79] |
Wriessnegger T, Augustin P, Engleder M, et al. Production of the sesquiterpenoid(+)-nootkatone by metabolic engineering of Pichia pastoris[J]. Metab Eng, 2014, 24: 18-29.
doi: 10.1016/j.ymben.2014.04.001 pmid: 24747046 |
| [80] |
Cha YP, Li W, Wu T, et al. Probing the synergistic ratio of P450/CPR to improve(+)-nootkatone production in Saccharomyces cerevisiae[J]. J Agric Food Chem, 2022, 70(3): 815-825.
doi: 10.1021/acs.jafc.1c07035 URL |
| [81] |
Ye Z, Huang Y, Shi B, et al. Coupling cell growth and biochemical pathway induction in Saccharomyces cerevisiae for production of(+)-valencene and its chemical conversion to(+)-nootkatone[J]. Metab Eng, 2022, 72: 107-115.
doi: 10.1016/j.ymben.2022.03.005 URL |
| [82] |
Zha WL, An TY, Li T, et al. Reconstruction of the biosynthetic pathway of santalols under control of the GAL regulatory system in yeast[J]. ACS Synth Biol, 2020, 9(2): 449-456.
doi: 10.1021/acssynbio.9b00479 pmid: 31940436 |
| [83] |
Diaz-Chavez ML, Moniodis J, Madilao LL, et al. Biosynthesis of sandalwood oil: Santalum album CYP76F cytochromes P450 produce santalols and bergamotol[J]. PLoS One, 2013, 8(9): e75053.
doi: 10.1371/journal.pone.0075053 URL |
| [84] |
Celedon JM, Chiang A, Yuen MMS, et al. Heartwood-specific transcriptome and metabolite signatures of tropical sandalwood(Santalum album)reveal the final step of(Z)-santalol fragrance biosynthesis[J]. Plant J, 2016, 86(4): 289-299.
doi: 10.1111/tpj.2016.86.issue-4 URL |
| [85] |
Jiang LH, Dong C, Liu TF, et al. Improved functional expression of cytochrome P450s in Saccharomyces cerevisiae through screening a cDNA library from Arabidopsis thaliana[J]. Front Bioeng Biotechnol, 2021, 9: 764851.
doi: 10.3389/fbioe.2021.764851 URL |
| [86] |
Zha WL, Zhang F, Shao JQ, et al. Rationally engineering santalene synthase to readjust the component ratio of sandalwood oil[J]. Nat Commun, 2022, 13(1): 2508.
doi: 10.1038/s41467-022-30294-8 pmid: 35523896 |
| [87] |
Eggersdorfer M, Laudert D, Létinois U, et al. One hundred years of vitamins-a success story of the natural sciences[J]. Angew Chem Int Ed Engl, 2012, 51(52): 12960-12990.
doi: 10.1002/anie.201205886 URL |
| [88] |
Aranda C, Municoy M, Guallar V, et al. Selective synthesis of 4-hydroxyisophorone and 4-ketoisophorone by fungal peroxygenases[J]. Catal Sci Technol, 2019, 9(6): 1398-1405.
doi: 10.1039/C8CY02114G URL |
| [89] |
Zhong W, Mao L, Xu Q, et al. Allylic oxidation of α-isophorone to keto-isophorone with molecular oxygen catalyzed by copper chloride in acetylacetone[J]. Appl Catal A Gen, 2014, 486: 193-200.
doi: 10.1016/j.apcata.2014.08.005 URL |
| [90] |
Hennig M, Püntener K, Scalone M. Synthesis of(R)- and(S)-4-hydroxyisophorone by ruthenium-catalyzed asymmetric transfer hydrogenation of ketoisophorone[J]. Tetrahedron Asymmetry, 2000, 11(9): 1849-1858.
doi: 10.1016/S0957-4166(00)00141-5 URL |
| [91] |
Kaluzna I, Schmitges T, Straatman H, et al. Enabling selective and sustainable P450 oxygenation technology. production of 4-hydroxy-α-isophorone on kilogram scale[J]. Org Process Res Dev, 2016, 20(4): 814-819.
doi: 10.1021/acs.oprd.5b00282 URL |
| [92] |
Tavanti M, Parmeggiani F, Castellanos JRG, et al. One-pot biocatalytic double oxidation of α-isophorone for the synthesis of ketoisophorone[J]. ChemCatChem, 2017, 9(17): 3338-3348.
doi: 10.1002/cctc.201700620 URL |
| [93] |
Dezvarei S, Lee JHZ, Bell SG. Stereoselective hydroxylation of isophorone by variants of the cytochromes P450 CYP102A1 and CYP101A1[J]. Enzyme Microb Technol, 2018, 111: 29-37.
doi: 10.1016/j.enzmictec.2018.01.002 URL |
| [94] |
Bell SG, Chen XH, Sowden RJ, et al. Molecular recognition in(+)-α-pinene oxidation by cytochrome P450cam[J]. J Am Chem Soc, 2003, 125(3): 705-714.
doi: 10.1021/ja028460a URL |
| [95] |
Meng SQ, Guo J, Nie KL, et al. Chemoenzymatic synthesis of fragrance compounds from stearic acid[J]. Chembiochem, 2019, 20(17): 2232-2235.
doi: 10.1002/cbic.201900210 pmid: 30983113 |
| [96] |
Manning J, Tavanti M, Porter JL, et al. Regio- and enantio-selective chemo-enzymatic C-H-lactonization of decanoic acid to(S)-δ-decalactone[J]. Angew Chem Int Ed Engl, 2019, 58(17): 5668-5671.
doi: 10.1002/anie.v58.17 URL |
| [1] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [2] | CHENG Ya-nan, ZHANG Wen-cong, ZHOU Yuan, SUN Xue, LI Yu, LI Qing-gang. Synthetic Pathway Construction of Producing 2'-fucosyllactose by Lactococcus lactis and Optimization of Fermentation Medium [J]. Biotechnology Bulletin, 2023, 39(9): 84-96. |
| [3] | ZHAO Si-jia, WANG Xiao-lu, SUN Ji-lu, TIAN Jian, ZHANG Jie. Modification of Pichia pastoris for Erythritol Production by Metabolic Engineering [J]. Biotechnology Bulletin, 2023, 39(8): 137-147. |
| [4] | YE Yun-fang, TIAN Qing-yin, SHI Ting-ting, WANG Liang, YUE Yuan-zheng, YANG Xiu-lian, WANG Liang-gui. Research Progress in the Biosynthesis and Regulation of β-ionone in Plants [J]. Biotechnology Bulletin, 2023, 39(8): 91-105. |
| [5] | WANG Ling, ZHUO Shen, FU Xue-sen, LIU Zi-xuan, LIU Xiao-rong, WANG Zhi-hui, ZHOU Ri-bao, LIU Xiang-dan. Advances in the Biosynthetic Pathways and Related Genes of Lotus Alkaloids [J]. Biotechnology Bulletin, 2023, 39(7): 56-66. |
| [6] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [7] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [8] | ZHOU Ding-ding, LI Hui-hu, TANG Xing-yong, YU Fa-xin, KONG Dan-yu, LIU Yi. Research Progress in the Biosynthesis and Regulation of Glycyrrhizic Acid and Liquiritin [J]. Biotechnology Bulletin, 2023, 39(5): 44-53. |
| [9] | WANG Mu-qiang, CHEN Qi, MA Wei, LI Chun-xiu, OUYANG Peng-fei, XU Jian-he. Advances in the Application of Machine Learning Methods for Directed Evolution of Enzymes [J]. Biotechnology Bulletin, 2023, 39(4): 38-48. |
| [10] | YAO Xiao-wen, LIANG Xiao, CHEN Qing, WU Chun-ling, LIU Ying, LIU Xiao-qiang, SHUI Jun, QIAO Yang, MAO Yi-ming, CHEN Yin-hua, ZHANG Yin-dong. Study on the Expression Pattern of Genes in Lignin Biosynthesis Pathway of Cassava Resisting to Tetranychus urticae [J]. Biotechnology Bulletin, 2023, 39(2): 161-171. |
| [11] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [12] | LI Yi-dan, SHAN Xiao-hui. Gibberellin Metabolism Regulation and Green Revolution [J]. Biotechnology Bulletin, 2022, 38(2): 195-204. |
| [13] | QIU Yi-bin, MA Yan-qin, SHA Yuan-yuan, ZHU Yi-fan, SU Er-zheng, LEI Peng, LI Sha, XU Hong. Research Progress in Molecular Genetic Manipulation Technology of Bacillus amyloliquefaciens and Its Application [J]. Biotechnology Bulletin, 2022, 38(2): 205-217. |
| [14] | MA Yan-qin, QIU Yi-bin, LI Sha, XU Hong. Research Progress in the Biosynthesis and Metabolic Engineering of Hyaluronic Acid [J]. Biotechnology Bulletin, 2022, 38(2): 252-262. |
| [15] | YANG Rui-xian, LIU Ping, WANG Zu-hua, RUAN Bao-shuo, WANG Zhi-da. Analysis of Antimicrobial Active Metabolites from Antagonistic Strains Against Fusarium solani [J]. Biotechnology Bulletin, 2022, 38(2): 57-66. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
