








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (2): 57-66.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0337
Previous Articles Next Articles
YANG Rui-xian( ), LIU Ping, WANG Zu-hua, RUAN Bao-shuo, WANG Zhi-da
), LIU Ping, WANG Zu-hua, RUAN Bao-shuo, WANG Zhi-da
Received:2021-03-18
Online:2022-02-26
Published:2022-03-09
YANG Rui-xian, LIU Ping, WANG Zu-hua, RUAN Bao-shuo, WANG Zhi-da. Analysis of Antimicrobial Active Metabolites from Antagonistic Strains Against Fusarium solani[J]. Biotechnology Bulletin, 2022, 38(2): 57-66.
| 序号 No. | 基因 Gene | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Product size/ bp | 产生的脂肽类物质 Lipopeptide |
|---|---|---|---|---|---|
| 1 | fenA | FenAa | AAGAGATTCAGTAAGTGGCCCATCCAG | 1 500 | Fengycin |
| FenAb | CGCCCTTTGGGAAGAGGTGC | ||||
| 2 | srfAA | Srfkn-1 | AGCCGTCCTGTCTGACGACG | 1 500 | Surfactin |
| Srfkn-2 | TCTGCTGCCATACCGCATAGTC | ||||
| 3 | ituD | ItuD-f ItuD-r | ATGAACAATCTTGCCTTTTTA TTATTTTAAATTCCGCAATT | 1 300 | Iturin |
Table 1 Primer sequence for lipopeptide biosynthesis genes fragments
| 序号 No. | 基因 Gene | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Product size/ bp | 产生的脂肽类物质 Lipopeptide |
|---|---|---|---|---|---|
| 1 | fenA | FenAa | AAGAGATTCAGTAAGTGGCCCATCCAG | 1 500 | Fengycin |
| FenAb | CGCCCTTTGGGAAGAGGTGC | ||||
| 2 | srfAA | Srfkn-1 | AGCCGTCCTGTCTGACGACG | 1 500 | Surfactin |
| Srfkn-2 | TCTGCTGCCATACCGCATAGTC | ||||
| 3 | ituD | ItuD-f ItuD-r | ATGAACAATCTTGCCTTTTTA TTATTTTAAATTCCGCAATT | 1 300 | Iturin |
| 序号 No. | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 基因 Gene |
|---|---|---|---|
| 1 | F1-F | CAGGTGCCGTTACAGGACAT | fenA |
| F2-R | TCGCAGGCTGAATCGCTTC | ||
| 2 | S1-F | CGGTCAGCAATACGGAAGTCT | srfAA |
| S2-R | TCAGTTCAGGACGGTTGTAGTAG | ||
| 3 | I1-F | ATTTCCTGGACAAGGGTCTCAAT | ituD |
| I2-R | TCGCATCGCTCGCTTCTTC | ||
| 4 | rpsJ-F | GAAACGGCAAAACGTTCTGG | rpsJ |
| rpsJ-R | GTGTTGGGTTCACAATGTCG |
Table 2 Primer sequences for lipopeptide biosynthesis genes in RT-qPCR
| 序号 No. | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 基因 Gene |
|---|---|---|---|
| 1 | F1-F | CAGGTGCCGTTACAGGACAT | fenA |
| F2-R | TCGCAGGCTGAATCGCTTC | ||
| 2 | S1-F | CGGTCAGCAATACGGAAGTCT | srfAA |
| S2-R | TCAGTTCAGGACGGTTGTAGTAG | ||
| 3 | I1-F | ATTTCCTGGACAAGGGTCTCAAT | ituD |
| I2-R | TCGCATCGCTCGCTTCTTC | ||
| 4 | rpsJ-F | GAAACGGCAAAACGTTCTGG | rpsJ |
| rpsJ-R | GTGTTGGGTTCACAATGTCG |
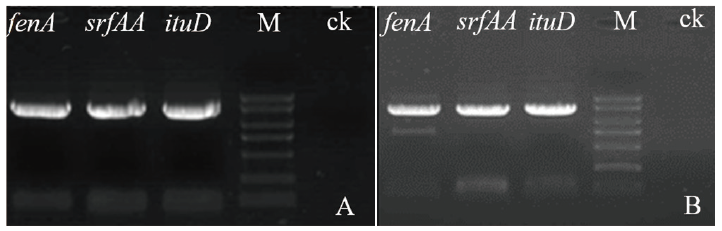
Fig. 3 Electrophoresis of PCR amplified lipopeptide biosy-nthesis gene fragments of strain md8 and md9 A:Amplified results of the lipopeptide biosynthesis gene fragments of md8. B:Amplified results of the lipopeptide biosynthesis gene fragments of md9. M:DL2000 marker
Fig.7 Inhibition effect of lipopeptide extract and separation components via gel chromatography of strain md8 and md9 inhibiting F. solani A:Inhibition effect of lipopeptide extract from strain md8 and md9. B:Plate inhibition effect of separation components of strain md8 via gel chromatography. C:Inhibition effect of separation components of strain md9 via gel chromatography
| [1] | 赵丹. 牡丹根部茎部真菌病害及病原鉴定[D]. 洛阳:河南科技大学, 2012. |
| Zhao D. The identification of pathogenic of peony root and stem fungal diseases[D]. Luoyang:Henan University of Science and Technology, 2012. | |
| [2] | 徐建强, 张吕醉, 等. 洛阳市牡丹病害种类及其症状识别[J]. 中国森林病虫, 2018, 37(1):35-38. |
| Xu JQ, Zhang LZ, et al. Peony diseases and their symptom identification in Luoyang[J]. For Pest Dis, 2018, 37(1):35-38. | |
| [3] | 成玉梅, 赵丹, 康业斌. 牡丹根腐病病原的形态与分子鉴定[J]. 北方园艺, 2015(16):116-119. |
| Cheng YM, Zhao D, Kang YB. Molecular and morphological identification of peony root rot pathogen[J]. North Hortic, 2015(16):116-119. | |
| [4] | 杨瑞先, 刘萍, 汪玉婷, 等. 洛阳地区牡丹根腐病病原菌的分离与鉴定[J]. 北方园艺, 2021(1):59-66. |
| Yang RX, Liu P, Wang YT, et al. Isolation and identification of root rot pathogens on Paeonia suffruticosa andr. in Luoyang[J]. North Hortic, 2021(1):59-66. | |
| [5] | 王桢, 贺春玲, 侯小改, 等. 6种杀菌剂对牡丹根腐病菌的室内毒力测定[J]. 安徽农业科学, 2015, 43(17):132-133, 137. |
| Wang Z, He CL, Hou XG, et al. Toxicity measurement of six fungicides against peony root rot disease[J]. J Anhui Agric Sci, 2015, 43(17):132-133, 137. | |
| [6] | 牛慧芳. 6种杀菌剂对油用牡丹根腐病的室内毒力及田间防效测定[J]. 内蒙古林业调查设计, 2017, 40(3):43-45. |
| Niu HF. Measurement of indoor toxicity and field efficacy of six fungicides against peony root rot[J]. Inn Mong For Investig Des, 2017, 40(3):43-45. | |
| [7] | 吴玉柱, 季延平, 刘慇, 等. 6种生防真菌、细菌防治牡丹根腐病的研究[J]. 山东林业科技, 2004, 34(6):39-40. |
| Wu YZ, Ji YP, Liu Y, et al. Control of tree peony root rot using 6 species of biocontrol fungi and bacteria[J]. J Shandong For Sci Technol, 2004, 34(6):39-40. | |
| [8] | 王雪山, 王善林, 杜秉海, 等. 牡丹根腐病拮抗菌的筛选与鉴定[J]. 山东农业科学, 2012, 44(7):90-94. |
| Wang XS, Wang SL, Du BH, et al. Screening and identification of antagonistic strains against Fusarium solani causing tree peony root rot[J]. Shandong Agric Sci, 2012, 44(7):90-94. | |
| [9] | 孟国庆, 张顺响, 黄一鸣, 等. 牡丹根腐病拮抗菌的分离与鉴定[J]. 菏泽学院学报, 2020, 42(5):106-110. |
| Meng GQ, Zhang SX, Huang YM, et al. Isolation and identification of antagonistic bacteria against root rot of peony[J]. J Heze Univ, 2020, 42(5):106-110. | |
| [10] | 杨瑞先, 姬俊华, 王祖华, 等. 牡丹根部内生细菌的分离鉴定及脂肽类物质的拮抗活性研究[J]. 微生物学通报, 2015, 42(6):1081-1088. |
| Yang RX, Ji JH, Wang ZH, et al. Isolation, indentification and inhibitory activity of lipopeptides of endophytic bacteria from the root of Paeonia suffruticosa[J]. Microbiol China, 2015, 42(6):1081-1088. | |
| [11] | 李全胜, 谢宗铭, 刘政, 等. 棉花黄萎病拮抗细菌H14的筛选鉴定及其拮抗机理分析[J]. 植物保护学报, 2018, 45(6):1204-1211. |
| Li QS, Xie ZM, Liu Z, et al. Screening and identification of antagonistic bacterium H14 against Verticillium dahliae Kleb. and its antagonistic mechanisms[J]. J Plant Prot, 2018, 45(6):1204-1211. | |
| [12] |
Koumoutsi A, Chen XH, Henne A, et al. Structural and functional characterization of gene clusters directing nonribosomal synjournal of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42[J]. J Bacteriol, 2004, 186(4):1084-1096.
doi: 10.1128/JB.186.4.1084-1096.2004 URL |
| [13] |
Hsieh FC, Lin TC, Meng M, et al. Comparing methods for identifying Bacillus strains capable of producing the antifungal lipopeptide iturin A[J]. Curr Microbiol, 2008, 56(1):1-5.
doi: 10.1007/s00284-007-9003-x URL |
| [14] |
Jordan S, Junker A, Helmann JD, et al. Regulation of LiaRS-dependent gene expression in Bacillus subtilis:identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system[J]. J Bacteriol, 2006, 188(14):5153-5166.
pmid: 16816187 |
| [15] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method[J]. Methods, 2001, 25(4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [16] | 桑建伟, 杨扬, 陈奕鹏, 等. 内生解淀粉芽孢杆菌BEB17脂肽类和聚酮类化合物的抑菌活性分析[J]. 植物病理学报, 2018, 48(3):402-412. |
| Sang JW, Yang Y, Chen YP, et al. Antibacterial activity analysis of lipopeptide and polyketide compounds produced by endophytic bacteria Bacillus amyloliquefaciens BEB17[J]. Acta Phytopathol Sin, 2018, 48(3):402-412. | |
| [17] | 邓建良. 解淀粉芽孢杆菌YN-1抑制植物病原真菌活性物质研究[D]. 武汉:华中农业大学, 2009. |
| Deng JL. Characterization of the antifungal substances produced by Bacillus amyloliquefaciens strain YN-1[D]. Wuhan:Huazhong Agricultural University, 2009. | |
| [18] | 李兴玉, 毛自朝, 吴毅歆, 等. 芽孢杆菌环脂肽类次生代谢产物的快速检测[J]. 植物病理学报, 2014, 44(6):718-722. |
| Li XY, Mao ZC, Wu YX, et al. Rapid detection of cyclic lipopeptide metabolites from Bacillus[J]. Acta Phytopathol Sin, 2014, 44(6):718-722. | |
| [19] |
Ongena M, Jacques P. Bacillus lipopeptides:versatile weapons for plant disease biocontrol[J]. Trends Microbiol, 2008, 16(3):115-125.
doi: 10.1016/j.tim.2007.12.009 pmid: 18289856 |
| [20] |
Shafi J, Tian H, Ji MS. Bacillus species as versatile weapons for plant pathogens:a review[J]. Biotechnol Biotechnol Equip, 2017, 31(3):446-459.
doi: 10.1080/13102818.2017.1286950 URL |
| [21] | 张学雯, 张学彬, 李红亚, 等. 黄曲霉拮抗菌Bacillus amyloliquefaciens B10-6-1产生的脂肽类抗生素的分离和鉴别[J]. 菌物学报, 2019, 38(4):594-602. |
| Zhang XW, Zhang XB, Li HY, et al. Isolation and identification of lipopeptide antibiotic produced by Bacillus amyloliquefaciens B10-6-1 antagonistic to Aspergillus flavus[J]. Mycosystema, 2019, 38(4):594-602. | |
| [22] |
Zhao P, Quan C, Jin L, et al. Sequence characterization and computational analysis of the non-ribosomal peptide synthetases controlling biosynjournal of lipopeptides, fengycins and bacillomycin D, from Bacillus amyloliquefaciens Q-426[J]. Biotechnol Lett, 2013, 35(12):2155-2163.
doi: 10.1007/s10529-013-1320-5 URL |
| [23] | 向亚萍, 周华飞, 刘永锋, 等. 解淀粉芽孢杆菌B1619脂肽类抗生素的分离鉴定及其对番茄枯萎病菌的抑制作用[J]. 中国农业科学, 2016, 49(15):2935-2944. |
| Xiang YP, Zhou HF, Liu YF, et al. Isolation and identification of lipopeptide antibiotics produced by Bacillus amyloliquefaciens B1619 and the inhibition of the lipopeptide antibiotics to Fusarium oxysporum f. sp. lycopersici[J]. Sci Agric Sin, 2016, 49(15):2935-2944. | |
| [24] | 杨洋. 解淀粉芽孢杆菌FZB42抗菌脂肽bacillomycinD对禾谷镰孢菌拮抗机制的研究[D]. 南京:南京农业大学, 2017. |
| Yang Y. Research about the antifungal mechanism of bacillomycinD from Bacillus amyloliquefaciens FZB42 Fusarium Graminiarum[D]. Nanjing:Nanjing Agricultural University, 2017. | |
| [25] | 曲晓旭, 刘洪霞, 高玲, 等. 芽孢杆菌产抗菌脂肽调控基因快速检测[J]. 南京农业大学学报, 2016, 39(5):858-864. |
| Qu XX, Liu HX, Gao L, et al. Rapid identification of the encoding genes of antimicrobial lipopeptides producted by Bacillus[J]. J Nanjing Agric Univ, 2016, 39(5):858-864. | |
| [26] | 朱洪磊, 马超, 等. 芽孢杆菌抗菌肽调控基因的PCR检测及其抑菌活性[J]. 山西农业科学, 2020, 48(3):304-308. |
| Zhu HL, Ma C, et al. PCR detection of regulation gene of antibacterial lipopeptide from Bacillus sp. and its antifungal activity[J]. J Shanxi Agric Sci, 2020, 48(3):304-308. | |
| [27] | Li B, Li Q, Xu Z, et al. Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soilborne fungal pathogens through the alteration of antifungal compounds production[J]. Front Microbiol, 2014, 5:636. |
| [28] |
Sajitha KL, Dev SA. Quantification of antifungal lipopeptide gene expression levels in Bacillus subtilis B1 during antagonism against sapstain fungus on rubberwood[J]. Biol Control, 2016, 96:78-85.
doi: 10.1016/j.biocontrol.2016.02.007 URL |
| [29] |
Sajitha KL, Dev SA, Maria Florence EJ. Identification and characterization of lipopeptides from Bacillus subtilis B1 against sapstain fungus of rubberwood through MALDI-TOF-MS and RT-PCR[J]. Curr Microbiol, 2016, 73(1):46-53.
doi: 10.1007/s00284-016-1025-9 pmid: 27004481 |
| [1] | YANG Dong-ya, QI Rui-xue LI, Zhao-xuan , LIN Wei, MA Hui, ZHANG Xue-yan. Screening, Identification and Growth-promoting Effect of Antagonistic Bacillus spp. Against Cucumber Fusarium solani [J]. Biotechnology Bulletin, 2023, 39(2): 211-220. |
| [2] | JIANG Jing-jing, ZHOU Zhao-xu, DU Hui, LYU Zhao-long, WANG Chun-ming, GUO Jian-guo, ZHANG Xin-rui, LI Ji-ping. Isolation and Identification of Apple Brown Rot Pathogen in Parts of Gansu and Screening of Antagonistic Bacteria [J]. Biotechnology Bulletin, 2023, 39(10): 209-218. |
| [3] | NIU Xin, ZHANG Ying, WANG Mao-jun, LIU Wen-long, LU Fu-ping, LI Yu. Effects of Different Integration Sites on the Expression of Exogenous Alkaline Protease in Bacillus amyloliquefaciens [J]. Biotechnology Bulletin, 2022, 38(4): 253-260. |
| [4] | QIU Yi-bin, MA Yan-qin, SHA Yuan-yuan, ZHU Yi-fan, SU Er-zheng, LEI Peng, LI Sha, XU Hong. Research Progress in Molecular Genetic Manipulation Technology of Bacillus amyloliquefaciens and Its Application [J]. Biotechnology Bulletin, 2022, 38(2): 205-217. |
| [5] | CAI Guo-lei, LU Xiao-kai, LOU Shui-zhu, YANG Hai-ying, DU Gang. Classification and Identification of Bacillus LM Based on Whole Genome and Study on Its Antibacterial Principle [J]. Biotechnology Bulletin, 2021, 37(8): 176-185. |
| [6] | LI Jin, PENG Ke-wei, PAN Qiu-yi, ZHU Zhe-yuan, PENG Di. Isolation and Identification of Bacillus amyloliquefaciens HR-2 and Biological Control of Rice Blast [J]. Biotechnology Bulletin, 2021, 37(3): 27-34. |
| [7] | LI Xin-yue, ZHANG Jin-fang, XU Xiao-jian, LU Fu-ping, LI Yu. Effects of Spore Formation Related Gene Deletion on Biomass and Extracellular Enzyme Expression of Bacillus amyloliquefaciens [J]. Biotechnology Bulletin, 2021, 37(3): 35-43. |
| [8] | YAO Ya-qian, CHENG Na-na, LI Pei-gen, LU Yi, WANG Yan-gang, LIN Rong-shan, ZHOU Bo, WANG Bing. Isolation and Identification of Bacillus amyloliquefaciens T-6 and Its Potential of Resisting Disease and Promoting Growth [J]. Biotechnology Bulletin, 2020, 36(9): 202-210. |
| [9] | LIN Jia-ming, GE Hui, LIN Ke-bing, YANG Zhang-wu, ZHOU Chen, WU Jian-shao, WANG Guo-dong, ZHANG Zhe, YANG Qiu-hua, WANG Yi-lei. Isolation,Identification and Antibiotic Sensitivity Analysis of Bacterial Pathogen from Litopenaeus vannamei with Black Gill Disease [J]. Biotechnology Bulletin, 2020, 36(8): 120-128. |
| [10] | GUO He-bao, WANG Xing, HE Shan-wen, ZHANG Xiao-xia. Phenotypic Characteristics Combined with Genomic Analysis to Identify Different Colony Morphology Bacillus velezensis ACCC 19742 [J]. Biotechnology Bulletin, 2020, 36(2): 142-148. |
| [11] | WANG Shi-wei, WANG Qing-hui. Research Advances in Functional Mechanisms of Bacillus amyloliquefacien [J]. Biotechnology Bulletin, 2020, 36(1): 150-159. |
| [12] | SONG Ben-chao, ZHAO Dong-mei, YANG Zhi-hui, ZHANG Dai, ZHAO Zhi, ZHU Jie-hua. Screening and Identification of an Antagonistic Bacterium Against Rhizoctonia solani and Analysis of Biocontrol Factor [J]. Biotechnology Bulletin, 2019, 35(8): 9-16. |
| [13] | LIU Duan-mu, WU Yi, LIU Yun, LIANG Zhi-hong. Screening,Identification and Antifungal Properties of a Bacterium with Antagonistic Activities Against Mycotoxin-producing Aspergillus spp. [J]. Biotechnology Bulletin, 2019, 35(8): 42-50. |
| [14] | LIU Chao, LIU Hong-wei, WANG Bu-qing, ZHAO Wen-ya, WANG Ya-na, ZHANG Li-ping. Isolation of Antifungal Substances from Bacillus amyloliquefaciens BA-26 and Its Antifungal Activity against Botrytis cinerea [J]. Biotechnology Bulletin, 2019, 35(7): 83-89. |
| [15] | LI Li, YAN Yue-gen, WU Hua-ming. Optimization of Shake Flask-fermentation Condition for Bacillus amyloliquefaciens M1 Strain and Its Denitrification Performance [J]. Biotechnology Bulletin, 2019, 35(5): 102-108. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||