








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (4): 59-70.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0971
Previous Articles Next Articles
WEI Ming WANG Xin-yu WU Guo-qiang ZHAO Meng( )
)
Received:2022-08-09
Online:2023-04-26
Published:2023-05-16
WEI Ming WANG Xin-yu WU Guo-qiang ZHAO Meng. The Role of NAD-dependent Deacetylase SRT in Plant Epigenetic Inheritance Regulation[J]. Biotechnology Bulletin, 2023, 39(4): 59-70.
| 物种 Species | 基因名称Gene name | 蛋白长度 Protein length/aa | 分子量 Mw/kD | 等电点 pI | 亚细胞定位 Subcellular localization | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 拟南芥Arabidopsis thaliana | AtSRT1 | 473 | 52.64 | 8.54 | Nucleus | [ |
| AtSRT2 | 376 | 41.87 | 9.08 | Mitochondria | ||
| 水稻Oryza sativa | OsSRT1 | 483 | 53.90 | 9.38 | Nucleus | [ |
| OsSRT2 | 393 | 43.47 | 8.79 | Mitochondria | ||
| 葡萄Vitis vinifera | VvSRT1 | 467 | 52.04 | 9.24 | / | [ |
| VvSRT2 | 382 | 41.99 | 9.09 | / | ||
| 番茄Solanum lycopersicum | SlSRT1 | 472 | 52.50 | 9.10 | Nucleus | [ |
| SlSRT2 | 389 | 43.06 | 8.97 | Mitochondria | ||
| 大豆Glycine max | GmSRT1 | 393 | 43.46 | 9.40 | Mitochondria chloroplast | [ |
| GmSRT2 | 392 | 43.21 | 9.32 | Mitochondria chloroplast | ||
| GmSRT3 | 434 | 48.21 | 9.11 | Nucleus chloroplast | ||
| GmSRT4 | 479 | 53.20 | 9.15 | Nucleus cytoplasm | ||
| 地钱Marchantia polymorpha | MpSIRT4 | 377 | 41.22 | 8.36 | Mitochondria | [ |
| MpSIRT5 | 285 | 30.75 | 7.02 | / | ||
| MpSIRT6 | 379 | 41.39 | 7.60 | Nucleus cytosol | ||
| 玉米Zea mays | ZmSRT1 | 437 | 49.05 | 9.31 | / | [ |
| ZmSRT2 | 351 | 39.19 | 8.91 | / | ||
| 茶树Camellia sinensis | CsSRT1 | 566 | 63.46 | 9.29 | Nucleus | [ |
| CsSRT2 | 460 | 51.36 | 8.82 | Chloroplast | ||
| CsSRT3 | 479 | 53.39 | 9.11 | Chloroplast | ||
| CsSRT4 | 177 | 19.77 | 9.58 | Nucleus | ||
| 石斛Dendrobium officinale | DoSRT1 | 409 | 45.50 | 8.19 | Nucleus | [ |
| DoSRT2 | 279 | 30.82 | 7.04 | Nucleus | ||
| 白梨Pyrus bretschneideri | PbSRT1 | 484 | 54.13 | 9.27 | Nucleus cytoplasm | [ |
| PbSRT2 | 394 | 43.46 | 8.80 | Chloroplast vacuole | ||
| 大麻Cannabis sativa | CsSRT1 | 484 | 53.61 | 9.04 | Nucleus chloroplast | [ |
| CsSRT2 | 397 | 43.49 | 8.37 | Nucleus chloroplast | ||
| 高粱Sorghum bicolor | SbSRT1 | 476 | 53.24 | 8.88 | Chloroplast | [ |
| SbSRT2 | 484 | 53.66 | 9.17 | Chloroplast | ||
| 甜菜Beta vulgaris | BvSRT1 | 496 | 55.64 | 9.22 | / | Unpublished data |
| BvSRT2 | 389 | 43.37 | 8.89 | / |
Table 1 SRT gene families in different plant species
| 物种 Species | 基因名称Gene name | 蛋白长度 Protein length/aa | 分子量 Mw/kD | 等电点 pI | 亚细胞定位 Subcellular localization | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 拟南芥Arabidopsis thaliana | AtSRT1 | 473 | 52.64 | 8.54 | Nucleus | [ |
| AtSRT2 | 376 | 41.87 | 9.08 | Mitochondria | ||
| 水稻Oryza sativa | OsSRT1 | 483 | 53.90 | 9.38 | Nucleus | [ |
| OsSRT2 | 393 | 43.47 | 8.79 | Mitochondria | ||
| 葡萄Vitis vinifera | VvSRT1 | 467 | 52.04 | 9.24 | / | [ |
| VvSRT2 | 382 | 41.99 | 9.09 | / | ||
| 番茄Solanum lycopersicum | SlSRT1 | 472 | 52.50 | 9.10 | Nucleus | [ |
| SlSRT2 | 389 | 43.06 | 8.97 | Mitochondria | ||
| 大豆Glycine max | GmSRT1 | 393 | 43.46 | 9.40 | Mitochondria chloroplast | [ |
| GmSRT2 | 392 | 43.21 | 9.32 | Mitochondria chloroplast | ||
| GmSRT3 | 434 | 48.21 | 9.11 | Nucleus chloroplast | ||
| GmSRT4 | 479 | 53.20 | 9.15 | Nucleus cytoplasm | ||
| 地钱Marchantia polymorpha | MpSIRT4 | 377 | 41.22 | 8.36 | Mitochondria | [ |
| MpSIRT5 | 285 | 30.75 | 7.02 | / | ||
| MpSIRT6 | 379 | 41.39 | 7.60 | Nucleus cytosol | ||
| 玉米Zea mays | ZmSRT1 | 437 | 49.05 | 9.31 | / | [ |
| ZmSRT2 | 351 | 39.19 | 8.91 | / | ||
| 茶树Camellia sinensis | CsSRT1 | 566 | 63.46 | 9.29 | Nucleus | [ |
| CsSRT2 | 460 | 51.36 | 8.82 | Chloroplast | ||
| CsSRT3 | 479 | 53.39 | 9.11 | Chloroplast | ||
| CsSRT4 | 177 | 19.77 | 9.58 | Nucleus | ||
| 石斛Dendrobium officinale | DoSRT1 | 409 | 45.50 | 8.19 | Nucleus | [ |
| DoSRT2 | 279 | 30.82 | 7.04 | Nucleus | ||
| 白梨Pyrus bretschneideri | PbSRT1 | 484 | 54.13 | 9.27 | Nucleus cytoplasm | [ |
| PbSRT2 | 394 | 43.46 | 8.80 | Chloroplast vacuole | ||
| 大麻Cannabis sativa | CsSRT1 | 484 | 53.61 | 9.04 | Nucleus chloroplast | [ |
| CsSRT2 | 397 | 43.49 | 8.37 | Nucleus chloroplast | ||
| 高粱Sorghum bicolor | SbSRT1 | 476 | 53.24 | 8.88 | Chloroplast | [ |
| SbSRT2 | 484 | 53.66 | 9.17 | Chloroplast | ||
| 甜菜Beta vulgaris | BvSRT1 | 496 | 55.64 | 9.22 | / | Unpublished data |
| BvSRT2 | 389 | 43.37 | 8.89 | / |
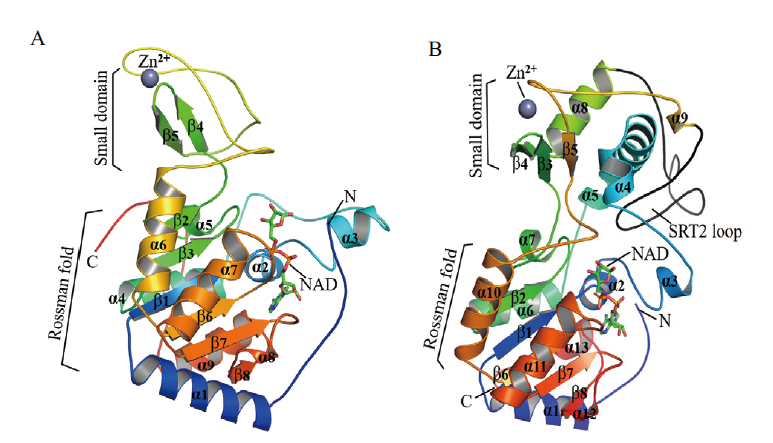
Fig. 1 Three-dimensional structure of SRTs in plants The three-dimensional structure of SIR2 domain in SRT1(A)and SRT2(B)proteins from Oryza sativa was predicted by homology modeling using SWISS-MODEL workspace[38]. The protein sequences of SRTs SIR2 domain were applied to search for a suitable template in the SWISS-MODEL Template Library(SMTL)and two crystal structures of SIR2(SRT1/2)from Homo sapiens(SMTL ID: 3k35.1 for SRT1)and Xenopus tropicalis(SMTL ID: 5oj7.1 for SRT2)were selected as the templates by the Global Model Quality Estimation(GMQE)values and the QMEANZ-scores[43-45]. Visualization and comparison of the SIR2 domains from SRT1 and SRT2 protein were performed through the Swiss PDB viewer and PyMOL program[46]

Fig. 2 Phylogenetic analysis of SRTs in different species SRTs基因的来源、名称及蛋白登录号如下: 拟南芥(Arabidopsis thaliana)AtSRT1(NP_200387.1), AtSRT2(NP_001078550.1);水稻(Oryza sativa)OsSRT1(XP_015636502.1), OsSRT2(XP_015618411.1);葡萄(Vitis riparia)VrSRT1(XP_034679101.1), VrSRT2(XP_010652926.1);番茄(Solanum lycopersicum)SlSRT1(XP_004244044.1), SlSRT2(XP_004236824.1);大豆(Glycine max)GmSRT1(XP_003522478.1), GmSRT1(XP_003528059.2), GmSRT3(KHN11152.1), GmSRT4(XP_003551434.1);地钱(Marchantia polymorpha)MpSIRT(PTQ32963.1), MpSIRT5(PTQ31328.1), MpSIRT6(PTQ40866.1);玉米(Zea mays)ZmSRT1(NP_001105577.1), ZmSRT2(XP_020400867.1);白梨(Pyrus bretschneideri)PbSRT1(XP_009375070.1), PbSRT2(XP_009375792.1);大麻(Cannabis sativa)CsSRT1(XP_030478610.1), CsSRT2(XP_030499415.1);高粱(Sorghum bicolor)SbSRT1(XP_021318881.1), SbSRT2(XP_002442918.2);甜菜(Beta vulgaris)BvSRT1(XP_019104724.1), BvSRT2(XP_010674411.1);菠菜(Spinacia oleracea)SoSRT1(XP_021842668.1), SoSRT2(XP_021835887.1);油菜(Brassica napus)BnSRT1(XP_022570011.1), BnSRT2(XP_013675778.1);芜菁(Brassica rapa)BrSRT1(XP_009132354.1), BrSRT2(XP_033140955.1);荠菜(Capsella rubella)CrSRT1(XP_023641545.1), CrSRT2(XP_023637530.1);烟草(Nicotiana tabacum)NtSRT1(XP_016490271.1), NtSRT2(XP_016482509.1);小麦(Triticum aestivum)TaSRT1(XP_044455034.1), TaSRT2(XP_044380222.1);谷子(Setaria italica)SiSRT1(XP_004975166.1), SiSRT2(XP_004977219.1);狗尾草(Setaria viridis)SvSRT1(XP_034603198.1), SvSRT2(XP_034606073.1);花生(Arachis hypogaea)AhSRT1(XP_025626741.1), AhSRT2(XP_025615786.1);木豆(Cajanus cajan)CcSRT1(XP_020232854.1), CcSRT2(XP_020234960.1);鹰嘴豆(Cicer arietinum)CaSRT1(XP_004516096.2), CaSRT2(XP_004502971.1);大叶栎(Quercus lobata)QlSRT1(XP_030931433.1), QlSRT2(XP_030940716.1);番木瓜(Carica papaya)CpSRT1(XP_021906115.1), CpSRT2(XP_021900464.1);南瓜(Cucurbita moschata)CmSRT1(XP_022936498.1), CmSRT2(XP_022948086.1);蒺藜苜蓿(Medicago truncatula)MtSRT1(XP_013460060.1), MtSRT2(XP_003602659.1);豇豆(Vigna unguiculata)VuSRT1(XP_027930984.1), VuSRT2(XP_027901777.1);冬瓜(Benincasa hispida)BhSRT1(XP_038900035.1), BhSRT1(XP_038902560.1);盐芥(Eutrema salsugineum)EsSRT1(XP_006401417.1), EsSRT2(XP_006399395.1)

Fig. 3 Diagram of histone acetylation/deacetylation modif-ication and dynamic regulation of gene transcription HAT:组蛋白乙酰转移酶 Histone acetyltransferase;HDACs:组蛋白去乙酰化酶 Histone deacetylases;Ac:乙酰基 Acetyl;TF:转录因子 Transcription factor
| [1] |
Luger K, Mäder AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution[J]. Nature, 1997, 389(6648): 251-260.
doi: 10.1038/38444 |
| [2] |
Zhou KD, Gaullier G, Luger K. Nucleosome structure and dynamics are coming of age[J]. Nat Struct Mol Biol, 2019, 26(1): 3-13.
doi: 10.1038/s41594-018-0166-x pmid: 30532059 |
| [3] |
Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications[J]. Cell Res, 2011, 21(3): 381-395.
doi: 10.1038/cr.2011.22 pmid: 21321607 |
| [4] |
Kouzarides T. Chromatin modifications and their function[J]. Cell, 2007, 128(4): 693-705.
doi: 10.1016/j.cell.2007.02.005 pmid: 17320507 |
| [5] |
Yang TT, Wang DY, Tian GM, et al. Chromatin remodeling complexes regulate genome architecture in Arabidopsis[J]. Plant Cell, 2022, 34(7): 2638-2651.
doi: 10.1093/plcell/koac117 URL |
| [6] |
Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease[J]. Nature, 2019, 571(7766): 489-499.
doi: 10.1038/s41586-019-1411-0 |
| [7] |
Ueda M, Seki M. Histone modifications form epigenetic regulatory networks to regulate abiotic stress response[J]. Plant Physiol, 2020, 182(1): 15-26.
doi: 10.1104/pp.19.00988 pmid: 31685643 |
| [8] |
Brukhin V, Albertini E. Epigenetic modifications in plant development and reproduction[J]. Epigenomes, 2021, 5(4): 25.
doi: 10.3390/epigenomes5040025 URL |
| [9] | Xia C, Tao Y, Li M, et al. Protein acetylation and deacetylation: an important regulatory modification in gene transcription(Review)[J]. Exp Ther Med, 2020, 20(4): 2923-2940. |
| [10] |
Wu X, Oh MH, Schwarz EM, et al. Lysine acetylation is a widespread protein modification for diverse proteins in Arabidop-sis[J]. Plant Physiol, 2011, 155(4): 1769-1778.
doi: 10.1104/pp.110.165852 URL |
| [11] |
Ma XJ, Lv SB, Zhang C, et al. Histone deacetylases and their functions in plants[J]. Plant Cell Rep, 2013, 32(4): 465-478.
doi: 10.1007/s00299-013-1393-6 pmid: 23408190 |
| [12] |
Chen XS, Ding AB, Zhong XH. Functions and mechanisms of plant histone deacetylases[J]. Sci China Life Sci, 2020, 63(2): 206-216.
doi: 10.1007/s11427-019-1587-x pmid: 31879846 |
| [13] |
Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis[J]. Curr Opin Genet Dev, 2001, 11(2): 155-161.
pmid: 11250138 |
| [14] |
Sadoul K, Boyault C, Pabion M, et al. Regulation of protein turnover by acetyltransferases and deacetylases[J]. Biochimie, 2008, 90(2): 306-312.
pmid: 17681659 |
| [15] |
van Gennip AH, Caron HN, et al. Histone deacetylases(HDACs): characterization of the classical HDAC family[J]. Biochem J, 2003, 370(Pt 3): 737-749.
doi: 10.1042/bj20021321 URL |
| [16] |
Tahir MS, Tian LN. HD2-type histone deacetylases: unique regulators of plant development and stress responses[J]. Plant Cell Rep, 2021, 40(9): 1603-1615.
doi: 10.1007/s00299-021-02688-3 pmid: 34041586 |
| [17] |
Hollender C, Liu ZC. Histone deacetylase genes in Arabidopsis development[J]. J Integr Plant Biol, 2008, 50(7): 875-885.
doi: 10.1111/j.1744-7909.2008.00704.x |
| [18] |
Yruela I, Moreno-Yruela C, Olsen CA. Zn2+-dependent histone deacetylases in plants: structure and evolution[J]. Trends Plant Sci, 2021, 26(7): 741-757.
doi: 10.1016/j.tplants.2020.12.011 URL |
| [19] |
Zheng WP. Review: the plant sirtuins[J]. Plant Sci, 2020, 293: 110434.
doi: 10.1016/j.plantsci.2020.110434 URL |
| [20] |
Brachmann CB, Sherman JM, Devine SE, et al. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability[J]. Genes Dev, 1995, 9(23): 2888-2902.
doi: 10.1101/gad.9.23.2888 URL |
| [21] |
Martínez-Redondo P, Vaquero A. The diversity of histone versus nonhistone sirtuin substrates[J]. Genes Cancer, 2013, 4(3/4): 148-163.
doi: 10.1177/1947601913483767 URL |
| [22] |
Pandey R, Müller A, Napoli CA, et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes[J]. Nucleic Acids Res, 2002, 30(23): 5036-5055.
doi: 10.1093/nar/gkf660 pmid: 12466527 |
| [23] |
Fu WQ, Wu KQ, Duan J. Sequence and expression analysis of histone deacetylases in rice[J]. Biochem Biophys Res Commun, 2007, 356(4): 843-850.
doi: 10.1016/j.bbrc.2007.03.010 URL |
| [24] | 符稳群, 吴克强, 张美, 等. 水稻组蛋白脱乙酰化酶SIR2的功能注释分析[J]. 安徽农业大学学报, 2008, 35(1): 70-75. |
| Fu WQ, Wu KQ, Zhang M, et al. Analysis of function notation of histone deacetylase SIR2 family in rice by bioinformatics strategy[J]. J Anhui Agric Univ, 2008, 35(1): 70-75. | |
| [25] |
Aquea F, Timmermann T, Arce-Johnson P. Analysis of histone acetyltransferase and deacetylase families of Vitis vinifera[J]. Plant Physiol Biochem, 2010, 48(2/3): 194-199.
doi: 10.1016/j.plaphy.2009.12.009 URL |
| [26] |
Aiese Cigliano R, Sanseverino W, Cremona G, et al. Genome-wide analysis of histone modifiers in tomato: Gaining an insight into their developmental roles[J]. BMC Genomics, 2013, 14: 57.
doi: 10.1186/1471-2164-14-57 pmid: 23356725 |
| [27] | Zhao LM, Lu JX, Zhang JX, et al. Identification and characterization of histone deacetylases in tomato(Solanum lycopersicum)[J]. Front Plant Sci, 2015, 5: 760. |
| [28] |
Yang C, Shen WJ, Chen HF, et al. Characterization and subcellular localization of histone deacetylases and their roles in response to abiotic stresses in soybean[J]. BMC Plant Biol, 2018, 18(1): 226.
doi: 10.1186/s12870-018-1454-7 pmid: 30305032 |
| [29] | 方淑梅, 冯乃杰, 梁喜龙. 大豆去乙酰化酶Sir2的生物信息学分析[J]. 贵州农业科学, 2015, 43(8): 34-37. |
| Fang SM, Feng NJ, Liang XL. Bioinformatics analysis of deacetylase Sir2 in soybean[J]. Guizhou Agric Sci, 2015, 43(8): 34-37. | |
| [30] |
Chu JS, Chen Z. Molecular identification of histone acetyltransferases and deacetylases in lower plant Marchantia polymorpha[J]. Plant Physiol Biochem, 2018, 132: 612-622.
doi: 10.1016/j.plaphy.2018.10.012 URL |
| [31] |
Zhang K, Yu L, Pang X, et al. In silico analysis of maize HDACs with an emphasis on their response to biotic and abiotic stresses[J]. PeerJ, 2020, 8: e8539.
doi: 10.7717/peerj.8539 URL |
| [32] |
Yuan LY, Dai HW, Zheng ST, et al. Genome-wide identification of the HDAC family proteins and functional characterization of CsHD2C, a HD2-type histone deacetylase gene in tea plant(Camellia sinensis L. O. Kuntze)[J]. Plant Physiol Biochem, 2020, 155: 898-913.
doi: 10.1016/j.plaphy.2020.07.047 URL |
| [33] |
Zhang MZ, Teixeira da Silva JA, Yu ZM, et al. Identification of histone deacetylase genes in Dendrobium officinale and their expression profiles under phytohormone and abiotic stress treatments[J]. PeerJ, 2020, 8: e10482.
doi: 10.7717/peerj.10482 URL |
| [34] |
Vall-Llaura N, Torres R, Lindo-García V, et al. PbSRT1 and PbSRT2 regulate pear growth and ripening yet displaying a species-specific regulation in comparison to other Rosaceae spp[J]. Plant Sci, 2021, 308: 110925.
doi: 10.1016/j.plantsci.2021.110925 URL |
| [35] |
Yang L, Meng XX, Chen SL, et al. Identification of the histone deacetylases gene family in hemp reveals genes regulating cannabinoids synthesis[J]. Front Plant Sci, 2021, 12: 755494.
doi: 10.3389/fpls.2021.755494 URL |
| [36] |
Du QL, Fang YP, Jiang JM, et al. Characterization of histone deacetylases and their roles in response to abiotic and PAMPs stresses in Sorghum bicolor[J]. BMC Genomics, 2022, 23(1): 28.
doi: 10.1186/s12864-021-08229-2 |
| [37] |
Blander G, Guarente L. The Sir2 family of protein deacetylases[J]. Annu Rev Biochem, 2004, 73: 417-435.
pmid: 15189148 |
| [38] |
Bordoli L, Kiefer F, Arnold K, et al. Protein structure homology modeling using SWISS-MODEL workspace[J]. Nat Protoc, 2009, 4(1): 1-13.
doi: 10.1038/nprot.2008.197 pmid: 19131951 |
| [39] |
Rossmann MG, Moras D, Olsen KW. Chemical and biological evolution of nucleotide-binding protein[J]. Nature, 1974, 250(463): 194-199.
doi: 10.1038/250194a0 |
| [40] |
Brzezinski K. S-adenosyl-l-homocysteine hydrolase: a structural perspective on the enzyme with two rossmann-fold domains[J]. Biomolecules, 2020, 10(12): 1682.
doi: 10.3390/biom10121682 URL |
| [41] |
Bellamacina CR. The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins[J]. FASEB J, 1996, 10(11): 1257-1269.
doi: 10.1096/fasebj.10.11.8836039 pmid: 8836039 |
| [42] |
Min J, Landry J, Sternglanz R, et al. Crystal structure of a SIR2 homolog-NAD complex[J]. Cell, 2001, 105(2): 269-279.
pmid: 11336676 |
| [43] |
Pannek M, Simic Z, Fuszard M, et al. Crystal structures of the mitochondrial deacylase Sirtuin 4 reveal isoform-specific acyl recognition and regulation features[J]. Nat Commun, 2017, 8(1): 1513.
doi: 10.1038/s41467-017-01701-2 pmid: 29138502 |
| [44] |
Jiménez-García B, Teixeira JMC, Trellet M, et al. PDB-tools web: a user-friendly interface for the manipulation of PDB files[J]. Proteins, 2021, 89(3): 330-335.
doi: 10.1002/prot.v89.3 URL |
| [45] |
Pan PW, Feldman JL, Devries MK, et al. Structure and biochemical functions of SIRT6[J]. J Biol Chem, 2011, 286(16): 14575-14587.
doi: 10.1074/jbc.M111.218990 pmid: 21362626 |
| [46] |
Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling[J]. Electrophoresis, 1997, 18(15): 2714-2723.
doi: 10.1002/elps.1150181505 pmid: 9504803 |
| [47] |
Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [48] |
Zhang H, Zhao Y, Zhou DX. Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes[J]. Nucleic Acids Res, 2017, 45(21): 12241-12255.
doi: 10.1093/nar/gkx825 pmid: 28981755 |
| [49] |
Liu XY, Wei W, Zhu WJ, et al. Histone deacetylase AtSRT1 links metabolic flux and stress response in Arabidopsis[J]. Mol Plant, 2017, 10(12): 1510-1522.
doi: 10.1016/j.molp.2017.10.010 URL |
| [50] |
König AC, Hartl M, Pham PA, et al. The Arabidopsis class II sirtuin is a lysine deacetylase and interacts with mitochondrial energy metabolism[J]. Plant Physiol, 2014, 164(3): 1401-1414.
doi: 10.1104/pp.113.232496 URL |
| [51] |
Chen B, Zang WW, Wang J, et al. The chemical biology of sirtuins[J]. Chem Soc Rev, 2015, 44(15): 5246-5264.
doi: 10.1039/c4cs00373j pmid: 25955411 |
| [52] |
Greiss S, Gartner A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation[J]. Mol Cells, 2009, 28(5): 407-415.
doi: 10.1007/s10059-009-0169-x pmid: 19936627 |
| [53] |
Chung PJ, Kim YS, Park SH, et al. Subcellular localization of rice histone deacetylases in organelles[J]. FEBS Lett, 2009, 583(13): 2249-2254.
doi: 10.1016/j.febslet.2009.06.003 pmid: 19505461 |
| [54] |
Borra MT, Langer MR, Slama JT, et al. Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases[J]. Biochemistry, 2004, 43(30): 9877-9887.
pmid: 15274642 |
| [55] |
Bheda P, Jing H, Wolberger C, et al. The substrate specificity of sirtuins[J]. Annu Rev Biochem, 2016, 85: 405-429.
doi: 10.1146/annurev-biochem-060815-014537 pmid: 27088879 |
| [56] |
Berger SL. The complex language of chromatin regulation during transcription[J]. Nature, 2007, 447(7143): 407-412.
doi: 10.1038/nature05915 |
| [57] |
Liu XC, Yang SG, Zhao ML, et al. Transcriptional repression by histone deacetylases in plants[J]. Mol Plant, 2014, 7(5): 764-772.
doi: 10.1093/mp/ssu033 pmid: 24658416 |
| [58] |
Imai S, Armstrong CM, Kaeberlein M, et al. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase[J]. Nature, 2000, 403(6771): 795-800.
doi: 10.1038/35001622 |
| [59] |
Vaquero A, Scher MB, Lee DH, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis[J]. Genes Dev, 2006, 20(10): 1256-1261.
doi: 10.1101/gad.1412706 URL |
| [60] |
North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases[J]. Genome Biol, 2004, 5(5): 224.
pmid: 15128440 |
| [61] |
Marmorstein R. Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases[J]. Biochem Soc Trans, 2004, 32(Pt 6): 904-909.
doi: 10.1042/BST0320904 URL |
| [62] |
Avalos JL, Boeke JD, Wolberger C. Structural basis for the mechanism and regulation of Sir2 enzymes[J]. Mol Cell, 2004, 13(5): 639-648.
pmid: 15023335 |
| [63] |
Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes[J]. Cold Spring Harb Perspect Biol, 2014, 6(4): a018713.
doi: 10.1101/cshperspect.a018713 URL |
| [64] |
Sauve AA, Celic I, Avalos J, et al. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions[J]. Biochemistry, 2001, 40(51): 15456-15463.
pmid: 11747420 |
| [65] |
Hawse WF, Wolberger C. Structure-based mechanism of ADP-ribosylation by sirtuins[J]. J Biol Chem, 2009, 284(48): 33654-33661.
doi: 10.1074/jbc.M109.024521 pmid: 19801667 |
| [66] |
Fahie K, Hu P, Swatkoski S, et al. Side chain specificity of ADP-ribosylation by a sirtuin[J]. FEBS J, 2009, 276(23): 7159-7176.
doi: 10.1111/j.1742-4658.2009.07427.x pmid: 19895577 |
| [67] | Wu LW, Ren DY, Hu SK, et al. Down-regulation of a nicotinate phosphoribosyltransferase gene, OsNaPRT1, leads to withered leaf tips[J]. Plant Physiol, 2016, 171(2): 1085-1098. |
| [68] |
Li Y, You L, Huang WF, et al. A FRET-based assay for screening SIRT6 modulators[J]. Eur J Med Chem, 2015, 96: 245-249.
doi: 10.1016/j.ejmech.2015.04.008 pmid: 25884115 |
| [69] |
Mautone N, Zwergel C, Mai A, et al. Sirtuin modulators: where are we now? A review of patents from 2015 to 2019[J]. Expert Opin Ther Pat, 2020, 30(6): 389-407.
doi: 10.1080/13543776.2020.1749264 pmid: 32228181 |
| [70] |
Alcaín FJ, Villalba JM. Sirtuin inhibitors[J]. Expert Opin Ther Pat, 2009, 19(3): 283-294.
doi: 10.1517/13543770902755111 pmid: 19441904 |
| [71] |
Bitterman KJ, Anderson RM, Cohen HY, et al. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1[J]. J Biol Chem, 2002, 277(47): 45099-45107.
doi: 10.1074/jbc.M205670200 pmid: 12297502 |
| [72] |
Bogan KL, Brenner C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition[J]. Annu Rev Nutr, 2008, 28: 115-130.
doi: 10.1146/nutr.2008.28.issue-1 URL |
| [73] |
Grubisha O, Smith BC, Denu JM. Small molecule regulation of Sir2 protein deacetylases[J]. FEBS J, 2005, 272(18): 4607-4616.
pmid: 16156783 |
| [74] |
Avalos JL, Celic I, Muhammad S, et al. Structure of a Sir2 enzyme bound to an acetylated p53 peptide[J]. Mol Cell, 2002, 10(3): 523-535.
pmid: 12408821 |
| [75] |
Singh S, Singh A, Yadav S, et al. Sirtinol, a Sir2 protein inhibitor, affects stem cell maintenance and root development in Arabidopsis thaliana by modulating auxin-cytokinin signaling components[J]. Sci Rep, 2017, 7: 42450.
doi: 10.1038/srep42450 |
| [76] |
Alcaín FJ, Villalba JM. Sirtuin activators[J]. Expert Opin Ther Pat, 2009, 19(4): 403-414.
doi: 10.1517/13543770902762893 pmid: 19441923 |
| [77] |
Sauve AA, Moir RD, Schramm VL, et al. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition[J]. Mol Cell, 2005, 17(4): 595-601.
pmid: 15721262 |
| [78] |
Kulkarni SS, Cantó C. The molecular targets of resveratrol[J]. Biochim Biophys Acta, 2015, 1852(6): 1114-1123.
doi: 10.1016/j.bbadis.2014.10.005 pmid: 25315298 |
| [79] |
Shakibaei M, Shayan P, Busch F, et al. Resveratrol mediated modulation of Sirt-1/Runx2 promotes osteogenic differentiation of mesenchymal stem cells: potential role of Runx2 deacetylation[J]. PLoS One, 2012, 7(4): e35712.
doi: 10.1371/journal.pone.0035712 URL |
| [80] |
Zhang F, Wang LK, Ko EE, et al. Histone deacetylases SRT1 and SRT2 interact with ENAP1 to mediate ethylene-induced transcriptional repression[J]. Plant Cell, 2018, 30(1): 153-166.
doi: 10.1105/tpc.17.00671 URL |
| [81] |
Fernie AR, Carrari F, Sweetlove LJ. Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport[J]. Curr Opin Plant Biol, 2004, 7(3): 254-261.
doi: 10.1016/j.pbi.2004.03.007 pmid: 15134745 |
| [82] |
Finkemeier I, Laxa M, Miguet L, et al. Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis[J]. Plant Physiol, 2011, 155(4): 1779-1790.
doi: 10.1104/pp.110.171595 pmid: 21311031 |
| [83] |
Cucurachi M, Busconi M, Morreale G, et al. Characterization and differential expression analysis of complete coding sequences of Vitis vinifera L. sirtuin genes[J]. Plant Physiol Biochem, 2012, 54: 123-132.
doi: 10.1016/j.plaphy.2012.02.017 URL |
| [84] |
Bond DM, Dennis ES, Pogson BJ, et al. Histone acetylation, vernalization insensitive 3, flowering locus c, and the vernalization response[J]. Mol Plant, 2009, 2(4): 724-737.
doi: S1674-2052(14)60755-3 pmid: 19825652 |
| [85] |
Zhang H, Lu Y, Zhao Y, et al. OsSRT1 is involved in rice seed development through regulation of starch metabolism gene expression[J]. Plant Sci, 2016, 248: 28-36.
doi: 10.1016/j.plantsci.2016.04.004 pmid: 27181944 |
| [86] |
Lee K, Park OS, Jung SJ, et al. Histone deacetylation-mediated cellular dedifferentiation in Arabidopsis[J]. J Plant Physiol, 2016, 191: 95-100.
doi: 10.1016/j.jplph.2015.12.006 URL |
| [87] |
Huang LM, Sun QW, Qin FJ, et al. Down-regulation of a silent information regulator2-related histone deacetylase gene, ossrt1, induces DNA fragmentation and cell death in rice[J]. Plant Physiol, 2007, 144(3): 1508-1519.
pmid: 17468215 |
| [88] |
Fang CY, Zhang H, Wan J, et al. Control of leaf senescence by an MeOH-jasmonates cascade that is epigenetically regulated by OsSRT1 in rice[J]. Mol Plant, 2016, 9(10): 1366-1378.
doi: 10.1016/j.molp.2016.07.007 URL |
| [89] |
Tang WS, Zhong L, Ding QQ, et al. Histone deacetylase AtSRT2 regulates salt tolerance during seed germination via repression of vesicle-associated membrane protein 714(VAMP714)in Arabidopsis[J]. New Phytol, 2022, 234(4): 1278-1293.
doi: 10.1111/nph.v234.4 URL |
| [90] |
Imran M, Shafiq S, Naeem MK, et al. Histone deacetylase(HDAC)gene family in allotetraploid cotton and its diploid progenitors: in silico identification, molecular characterization, and gene expression analysis under multiple abiotic stresses, DNA damage and phytohormone treatments[J]. Int J Mol Sci, 2020, 21(1): 321.
doi: 10.3390/ijms21010321 URL |
| [91] |
Wang CZ, Gao F, Wu JG, et al. Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression[J]. Plant Cell Physiol, 2010, 51(8): 1291-1299.
doi: 10.1093/pcp/pcq087 URL |
| [92] |
Zhang F, Wang LK, Qi B, et al. EIN2 mediates direct regulation of histone acetylation in the ethylene response[J]. Proc Natl Acad Sci USA, 2017, 114(38): 10274-10279.
doi: 10.1073/pnas.1707937114 pmid: 28874528 |
| [93] |
Zhao H, Yin CC, Ma B, et al. Ethylene signaling in rice and Arabidopsis: new regulators and mechanisms[J]. J Integr Plant Biol, 2021, 63(1): 102-125.
doi: 10.1111/jipb.v63.1 URL |
| [94] |
Huang CRL, Burns KH, Boeke JD. Active transposition in genomes[J]. Annu Rev Genet, 2012, 46: 651-675.
doi: 10.1146/annurev-genet-110711-155616 pmid: 23145912 |
| [95] |
Zhong XC, Zhang H, Zhao Y, et al. The rice NAD+-dependent histone deacetylase OsSRT1 targets preferentially to stress- and metabolism-related genes and transposable elements[J]. PLoS One, 2013, 8(6): e66807.
doi: 10.1371/journal.pone.0066807 URL |
| [1] | HU Hai-lin, XU Li, LI Xiao-xu, WANG Chen-can, MEI Man, DING Wen-jing, ZHAO Yuan-yuan. Advances in the Regulation of Plant Growth, Development and Stress Physiology by Small Peptide Hormones [J]. Biotechnology Bulletin, 2023, 39(7): 13-25. |
| [2] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [3] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [4] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [5] | REN Pei-dong, PENG Jian-ling, LIU Sheng-hang, YAO Zi-ting, ZHU Gui-ning, LU Guang-tao, LI Rui-fang. Isolation and Identification of a Bacillus safensis Strain GX-H6 and Its Biocontrol Effect on Bacterial Leaf Streak of Rice [J]. Biotechnology Bulletin, 2023, 39(5): 243-253. |
| [6] | XUE Jiao ZHU Qing-feng FENG Yan-zhao CHEN Pei LIU Wen-hua ZHANG Ai-xia LIU Qin-jian ZHANG Qi YU Yang. Advances in Upstream Open Reading Frame in Plant Genes [J]. Biotechnology Bulletin, 2023, 39(4): 157-165. |
| [7] | SANG Tian, WANG Peng-cheng. Research Progress in Plant SUMOylation [J]. Biotechnology Bulletin, 2023, 39(3): 1-12. |
| [8] | YAN Xiong-ying, WANG Zhen, WANG Xia, YANG Shi-hui. Microbial Sulfur Metabolism and Stress Resistance [J]. Biotechnology Bulletin, 2023, 39(11): 150-167. |
| [9] | ZHANG Hong-hong, FANG Xiao-feng. Advances in the Regulation of Stress Sensing and Responses by Phase Separation in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 44-53. |
| [10] | SUN Yu-tong, LIU De-shuai, QI Xun, FENG Mei, HUANG Xu-zheng, YAO Wen-kong. Advances in Jasmonic Acid Regulating Plant Growth and Development as Well as Stress [J]. Biotechnology Bulletin, 2023, 39(11): 99-109. |
| [11] | LIU Yuan-yuan, WEI Chuan-zheng, XIE Yong-bo, TONG Zong-jun, HAN Xing, GAN Bing-cheng, XIE Bao-gui, YAN Jun-jie. Characteristics of Class II Peroxidase Gene Expression During Fruiting Body Development and Stress Response in Flammulina filiformis [J]. Biotechnology Bulletin, 2023, 39(11): 340-349. |
| [12] | AN Chang, LU Lin, SHEN Meng-qian, CHEN Sheng-zhen, YE Kang-zhuo, QIN Yuan, ZHENG Ping. Research Progress of bHLH Gene Family in Plants and Its Application Prospects in Medical Plants [J]. Biotechnology Bulletin, 2023, 39(10): 1-16. |
| [13] | LI Jian-jian, HE Chen-jing, HUANG Xiao-ping, XIANG Tai-he. Research Progress in the Regulation of Development and Stress Response by Long Non-coding RNAs in Plants [J]. Biotechnology Bulletin, 2023, 39(1): 48-58. |
| [14] | WANG Nan-nan, WANG Wen-jia, ZHU Qiang. Research Progress of microRNAs in Plant Stress Responses [J]. Biotechnology Bulletin, 2022, 38(8): 1-11. |
| [15] | XUE Man-de, ZHAO Feng-yue, LI Jie, JIANG Dan-hua. Advances in Histone Variants in Plant Epigenetic Regulation [J]. Biotechnology Bulletin, 2022, 38(7): 1-12. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||