








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (8): 159-164.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0173
Previous Articles Next Articles
YANG Yu-mei1,2( ), ZHANG Kun-xiao1,2(
), ZHANG Kun-xiao1,2( )
)
Received:2023-03-02
Online:2023-08-26
Published:2023-09-05
Contact:
ZHANG Kun-xiao
E-mail:yangyumei03@163.com;2015000022@jou.edu.cn
YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology[J]. Biotechnology Bulletin, 2023, 39(8): 159-164.
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| sgRNA-AAVS1-h1-top | CACCGCGTGGGTTTATCAACCACTT |
| sgRNA-AAVS1-h1-bot | AAACAAGTGGTTGATAAACCCACGC |
| sgRNA-AAVS1-h2-top | CACCGCACGTGGGTTTATCAACCAC |
| sgRNA-AAVS1-h2-bot | AAACGTGGTTGATAAACCCACGTGC |
| sgRNA-AAVS1-h3-top | CACCGACGTGGGTTTATCAACCACT |
| sgRNA-AAVS1-h3-bot | AAACAGTGGTTGATAAACCCACGTC |
| PUC-AAVS1-F | ACCATGATTACGCCAAGCTTGTGTTCACCAGGTCGTGGC |
| PUC-AAVS1-R | TGTACTGAGAGTGCACCATATGGACCTGAACTGGAGCTGAGG |
Table 1 PC construction of sgRNA expression vector primers and genome amplification primer information
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| sgRNA-AAVS1-h1-top | CACCGCGTGGGTTTATCAACCACTT |
| sgRNA-AAVS1-h1-bot | AAACAAGTGGTTGATAAACCCACGC |
| sgRNA-AAVS1-h2-top | CACCGCACGTGGGTTTATCAACCAC |
| sgRNA-AAVS1-h2-bot | AAACGTGGTTGATAAACCCACGTGC |
| sgRNA-AAVS1-h3-top | CACCGACGTGGGTTTATCAACCACT |
| sgRNA-AAVS1-h3-bot | AAACAGTGGTTGATAAACCCACGTC |
| PUC-AAVS1-F | ACCATGATTACGCCAAGCTTGTGTTCACCAGGTCGTGGC |
| PUC-AAVS1-R | TGTACTGAGAGTGCACCATATGGACCTGAACTGGAGCTGAGG |

Fig. 2 Design schematic diagram of ERK-SPARK knock-in plasmid AAVS1-HA-L/AAVS1-HA-R: Homology arms in AAS1 locus; SA: splicing acceptor; T2A: 2A peptide; PuroR: puromycin N-acetyltransferase; rtTA: improved tetracycline-controlled transactivator; poly A: polyadenylation signal; Tet-on: Tet-responsive element
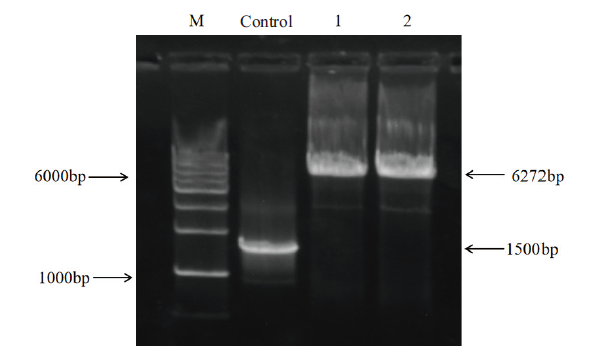
Fig. 3 Genomic PCR detection of the ERK-SPARK expression at the AAVS1 locus in KYSE-150 cells M: DNA marker; WT: KYSE-150 cells wild type; 1: ERK-SPARK knock-in cells;2: ERK-SPARK-mutation knock-in cells
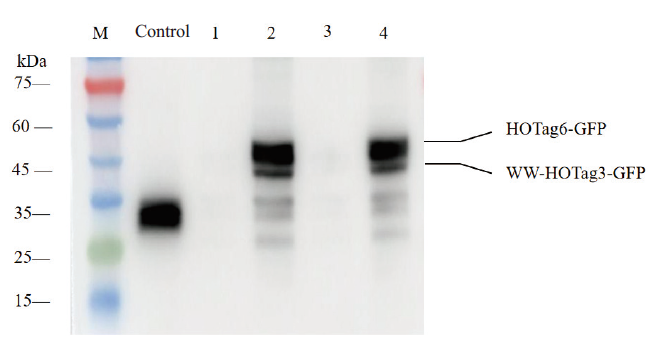
Fig. 4 Validating the doxycycline-induced expression of the ERK-SPARK reporter in KYSE-150 cells by Western blot M: Protein marker; control: anti-GFP; l: anti-GFP, ERK-SPARK knock-in cells, Dox(-); 2: anti-GFP, ERK-SPARK knock-in cells, Dox(+); 3: anti-GFP, ERK-SPARK-mutation knock-in cells, Dox(-); 4: anti-GFP, ERK-SPARK-mutation knock-in cells, Dox(+)
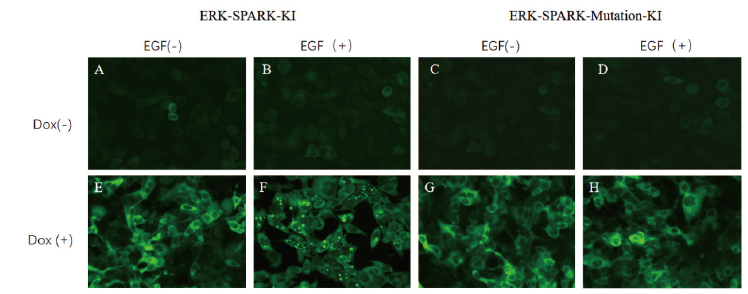
Fig. 5 Fluorescence images of ERK-SPARK knock-in cells stimulated with EGF(20X) A: Dox(-)EGF(-); B: Dox(-)EGF(+); C: Dox(-)EGF(-); D: Dox(-)EGF(+); E: Dox(+)EGF(-); F: Dox(+)EGF(+); G: Dox(+)EGF(-); H: Dox(+)EGF(+)
| [1] |
Li PL, Banjade S, Cheng HC, et al. Phase transitions in the assembly of multivalent signalling proteins[J]. Nature, 2012, 483(7389): 336-340.
doi: 10.1038/nature10879 |
| [2] |
Zhang Q, Huang H, Zhang LQ, et al. Visualizing dynamics of cell signaling in vivo with a phase separation-based kinase reporter[J]. Mol Cell, 2018, 69(2): 334-346.e4.
doi: 10.1016/j.molcel.2017.12.008 URL |
| [3] |
Romei MG, Boxer SG. Split green fluorescent proteins: scope, limitations, and outlook[J]. Annu Rev Biophys, 2019, 48(1): 19-44.
doi: 10.1146/biophys.2019.48.issue-1 URL |
| [4] |
Tebo AG, Gautier A. A split fluorescent reporter with rapid and reversible complementation[J]. Nat Commun, 2019, 10(1): 2822.
doi: 10.1038/s41467-019-10855-0 pmid: 31249300 |
| [5] |
Liu WF, Deng MY, Yang CM, et al. Genetically encoded single circularly permuted fluorescent protein-based intensity indicators[J]. J Phys D Appl Phys, 2019, 53(11): 113001.
doi: 10.1088/1361-6463/ab5dd8 URL |
| [6] |
Wu TC, Pang Y, Ai HW. Circularly permuted far-red fluorescent proteins[J]. Biosensors, 2021, 11(11): 438.
doi: 10.3390/bios11110438 URL |
| [7] |
Deng HT, Li JY, Zhou Y, et al. Genetic engineering of circularly permuted yellow fluorescent protein reveals intracellular acidification in response to nitric oxide stimuli[J]. Redox Biol, 2021, 41: 101943.
doi: 10.1016/j.redox.2021.101943 URL |
| [8] |
Li L, Cheng YC, Shen SY, et al. Sensitive detection via the time-resolved fluorescence of circularly permuted yellow fluorescent protein biosensors[J]. Sens Actuat B Chem, 2020, 321: 128614.
doi: 10.1016/j.snb.2020.128614 URL |
| [9] |
Kobayashi H, Picard LP, Schönegge AM, et al. Bioluminescence resonance energy transfer-based imaging of protein-protein interactions in living cells[J]. Nat Protoc, 2019, 14(4): 1084-1107.
doi: 10.1038/s41596-019-0129-7 pmid: 30911173 |
| [10] |
Zhang XJ, Hu Y, Yang XT, et al. FÖrster resonance energy transfer(FRET)-based biosensors for biological applications[J]. Biosens Bioelectron, 2019, 138: 111314.
doi: 10.1016/j.bios.2019.05.019 URL |
| [11] |
Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology[J]. Annu Rev Cell Dev Biol, 2014, 30: 39-58.
doi: 10.1146/annurev-cellbio-100913-013325 pmid: 25288112 |
| [12] |
Woolfson DN, Bartlett GJ, Burton AJ, et al. De novo protein design: how do we expand into the universe of possible protein structures?[J]. Curr Opin Struct Biol, 2015, 33: 16-26.
doi: 10.1016/j.sbi.2015.05.009 URL |
| [13] |
Grigoryan G, Kim YH, Acharya R, et al. Computational design of virus-like protein assemblies on carbon nanotube surfaces[J]. Science, 2011, 332(6033): 1071-1076.
doi: 10.1126/science.1198841 pmid: 21617073 |
| [14] |
Huang PS, Oberdorfer G, Xu CF, et al. High thermodynamic stability of parametrically designed helical bundles[J]. Science, 2014, 346(6208): 481-485.
doi: 10.1126/science.1257481 URL |
| [15] |
Thomson AR, Wood CW, Burton AJ, et al. Computational design of water-soluble α-helical barrels[J]. Science, 2014, 346(6208): 485-488.
doi: 10.1126/science.1257452 pmid: 25342807 |
| [16] |
Sheridan DL, Kong Y, Parker SA, et al. Substrate discrimination among mitogen-activated protein kinases through distinct docking sequence motifs[J]. J Biol Chem, 2008, 283(28): 19511-19520.
doi: 10.1074/jbc.M801074200 pmid: 18482985 |
| [17] |
Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823.
doi: 10.1126/science.1231143 pmid: 23287718 |
| [18] |
Koch B, Nijmeijer B, Kueblbeck M, et al. Generation and validation of homozygous fluorescent knock-in cells using CRISPR-Cas9 genome editing[J]. Nat Protoc, 2018, 13(6): 1465-1487.
doi: 10.1038/nprot.2018.042 pmid: 29844520 |
| [19] |
Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096.
doi: 10.1126/science.1258096 URL |
| [20] |
Kelly JJ, Saee-Marand M, Nyström NN, et al. Safe Harbor-targeted CRISPR-Cas9 homology-independent targeted integration for multimodality reporter gene-based cell tracking[J]. Sci Adv, 2021, 7(4): eabc3791.
doi: 10.1126/sciadv.abc3791 URL |
| [1] | CHENG Ya-nan, ZHANG Wen-cong, ZHOU Yuan, SUN Xue, LI Yu, LI Qing-gang. Synthetic Pathway Construction of Producing 2'-fucosyllactose by Lactococcus lactis and Optimization of Fermentation Medium [J]. Biotechnology Bulletin, 2023, 39(9): 84-96. |
| [2] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [3] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [4] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [5] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [6] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [7] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [8] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [9] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [10] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [11] | Olalekan Amoo, HU Li-min, ZHAI Yun-gu, FAN Chu-chuan, ZHOU Yong-ming. Regulation of Shoot Branching by BRANCHED1 in Brassica napus Based on Gene Editing Technology [J]. Biotechnology Bulletin, 2022, 38(4): 97-105. |
| [12] | DING Ya-qun, DING Ning, XIE Shen-min, HUANG Meng-na, ZHANG Yu, ZHANG Qin, JIANG Li. Construction of Vps28 Knock-out Mice and Model Study of the Impact on Lactation and Immune Traits [J]. Biotechnology Bulletin, 2022, 38(3): 164-172. |
| [13] | YAN Jiong, FENG Chen-yi, GAO Xue-kun, XU Xiang, YANG Jia-min, CHEN Zhao-yang. Construction of Homozygous Plin1-knockout Mouse Model and Phenotype Analysis Based on CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2022, 38(3): 173-180. |
| [14] | ZHONG Jing, SUN Ling-ling, ZHANG Shu, MENG Yuan, ZHI Yi-fei, TU Li-qing, XU Tian-peng, PU Li-ping, LU Yang-qing. Effect of Knocking Out the Mda5 Gene by CRISPR/Cas9 Technology on the Replication of Newcastle Disease and Infectious Bursal Virus [J]. Biotechnology Bulletin, 2022, 38(11): 90-96. |
| [15] | ZONG Mei, HAN Shuo, GUO Ning, DUAN Meng-meng, LIU Fan, WANG Gui-xiang. Production of Marker-free Mutants of Brassica campestris Mediated by CRISPR/Cas9 Through Vacuum Infiltration [J]. Biotechnology Bulletin, 2022, 38(10): 159-163. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||